An Affinity Propagation-Based DNA Motif Discovery Algorithm
- PMID: 26347887
- PMCID: PMC4547008
- DOI: 10.1155/2015/853461
An Affinity Propagation-Based DNA Motif Discovery Algorithm
Abstract
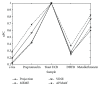
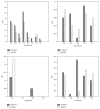
The planted (l, d) motif search (PMS) is one of the fundamental problems in bioinformatics, which plays an important role in locating transcription factor binding sites (TFBSs) in DNA sequences. Nowadays, identifying weak motifs and reducing the effect of local optimum are still important but challenging tasks for motif discovery. To solve the tasks, we propose a new algorithm, APMotif, which first applies the Affinity Propagation (AP) clustering in DNA sequences to produce informative and good candidate motifs and then employs Expectation Maximization (EM) refinement to obtain the optimal motifs from the candidate motifs. Experimental results both on simulated data sets and real biological data sets show that APMotif usually outperforms four other widely used algorithms in terms of high prediction accuracy.
Figures
Similar articles
-
PairMotif: A new pattern-driven algorithm for planted (l, d) DNA motif search.PLoS One. 2012;7(10):e48442. doi: 10.1371/journal.pone.0048442. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23119020 Free PMC article.
-
A cluster refinement algorithm for motif discovery.IEEE/ACM Trans Comput Biol Bioinform. 2010 Oct-Dec;7(4):654-68. doi: 10.1109/TCBB.2009.25. IEEE/ACM Trans Comput Biol Bioinform. 2010. PMID: 21030733
-
An Efficient Exact Algorithm for Planted Motif Search on Large DNA Sequence Datasets.IEEE/ACM Trans Comput Biol Bioinform. 2024 Sep-Oct;21(5):1542-1551. doi: 10.1109/TCBB.2024.3404136. Epub 2024 Oct 9. IEEE/ACM Trans Comput Biol Bioinform. 2024. PMID: 38801693
-
An extension and novel solution to the (l,d)-motif challenge problem.Genome Inform. 2004;15(2):63-71. Genome Inform. 2004. PMID: 15706492 Review.
-
A Review on Planted (l, d) Motif Discovery Algorithms for Medical Diagnose.Sensors (Basel). 2022 Feb 5;22(3):1204. doi: 10.3390/s22031204. Sensors (Basel). 2022. PMID: 35161949 Free PMC article. Review.
Cited by
-
A Clustering Approach for Motif Discovery in ChIP-Seq Dataset.Entropy (Basel). 2019 Aug 16;21(8):802. doi: 10.3390/e21080802. Entropy (Basel). 2019. PMID: 33267515 Free PMC article.
-
Review of Different Sequence Motif Finding Algorithms.Avicenna J Med Biotechnol. 2019 Apr-Jun;11(2):130-148. Avicenna J Med Biotechnol. 2019. PMID: 31057715 Free PMC article. Review.
-
An Entropy-Based Position Projection Algorithm for Motif Discovery.Biomed Res Int. 2016;2016:9127474. doi: 10.1155/2016/9127474. Epub 2016 Nov 2. Biomed Res Int. 2016. PMID: 27882329 Free PMC article.
References
-
- Pevzner P. A., Sze S.-H. Combinatorial approaches to finding subtle signals in DNA sequences. Proceedings of the 8th International Conference on Intelligent Systems for Molecular Biology; August 2000; San Diego, Calif, USA. pp. 269–278. - PubMed
-
- Chin F. Y. L., Leung H. C. M. Voting algorithms for discovering long motifs. Proceedings of the 3rd Asia-Pacific Bioinformatics Conference (APBC '05); January 2005; pp. 261–271.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources