Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPα
- PMID: 26348894
- PMCID: PMC4607129
- DOI: 10.1172/JCI77095
Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPα
Abstract
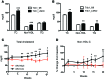
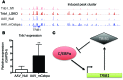
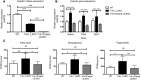
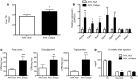
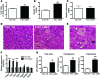
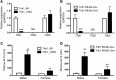
Variants near the gene TRIB1 are significantly associated with several plasma lipid traits, circulating liver enzymes, and the development of coronary artery disease in humans; however, it is not clear how its protein product tribbles-1 regulates lipid metabolism. Here, we evaluated mice harboring a liver-specific deletion of Trib1 (Trib1_LSKO) to elucidate the role of tribbles-1 in mammalian hepatic lipid metabolism. These mice exhibited increased hepatic triglyceride (TG) content, lipogenic gene transcription, and de novo lipogenesis. Microarray analysis revealed altered transcription of genes that are downstream of the transcription factor C/EBPα, and Trib1_LSKO mice had increased hepatic C/EBPα protein. Hepatic overexpression of C/EBPα in WT mice phenocopied Trib1_LSKO livers, and hepatic knockout of Cebpa in Trib1_LSKO mice revealed that C/EBPα is required for the increased lipogenesis. Using ChIP-Seq, we found that Trib1_LSKO mice had increased DNA-bound C/EBPα near lipogenic genes and the Trib1 gene, which itself was transcriptionally upregulated by C/EBPα overexpression. Together, our results reveal that tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPα, which in turn transcriptionally upregulates Trib1. These data suggest an important role for C/EBPα in mediating the lipogenic effects of hepatic Trib1 deletion and provide insight into the association between TRIB1 and plasma lipids, and liver traits in humans.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

