Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment
- PMID: 26348897
- PMCID: PMC4607132
- DOI: 10.1172/JCI77995
Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment
Abstract
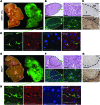
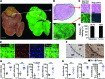
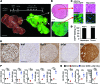
In many organs, including the intestine and skin, cancers originate from cells of the stem or progenitor compartment. Despite its nomenclature, the cellular origin of hepatocellular carcinoma (HCC) remains elusive. In contrast to most organs, the liver lacks a defined stem cell population for organ maintenance. Previous studies suggest that both hepatocytes and facultative progenitor cells within the biliary compartment are capable of generating HCC. As HCCs with a progenitor signature carry a worse prognosis, understanding the origin of HCC is of clinical relevance. Here, we used complementary fate-tracing approaches to label the progenitor/biliary compartment and hepatocytes in murine hepatocarcinogenesis. In genotoxic and genetic models, HCCs arose exclusively from hepatocytes but never from the progenitor/biliary compartment. Cytokeratin 19-, A6- and α-fetoprotein-positive cells within tumors were hepatocyte derived. In summary, hepatocytes represent the cell of origin for HCC in mice, and a progenitor signature does not reflect progenitor origin, but dedifferentiation of hepatocyte-derived tumor cells.
Figures



Similar articles
-
Hepatic progenitor cells, stem cells, and AFP expression in models of liver injury.Int J Exp Pathol. 2006 Oct;87(5):343-59. doi: 10.1111/j.1365-2613.2006.00485.x. Int J Exp Pathol. 2006. PMID: 16965562 Free PMC article.
-
Different lineages of chemically induced hepatocellular carcinoma in rats defined by monoclonal antibodies.Cancer Res. 1989 Sep 1;49(17):4894-900. Cancer Res. 1989. PMID: 2474377
-
Gankyrin-mediated dedifferentiation facilitates the tumorigenicity of rat hepatocytes and hepatoma cells.Hepatology. 2011 Oct;54(4):1259-72. doi: 10.1002/hep.24530. Hepatology. 2011. PMID: 21735473
-
Multistage rodent hepatocarcinogenesis.Prog Liver Dis. 1986;8:597-620. Prog Liver Dis. 1986. PMID: 3520671 Review. No abstract available.
-
Cellular origin of cancer: dedifferentiation or stem cell maturation arrest?Environ Health Perspect. 1993 Dec;101 Suppl 5(Suppl 5):15-26. doi: 10.1289/ehp.93101s515. Environ Health Perspect. 1993. PMID: 7516873 Free PMC article. Review.
Cited by
-
Adeno-Associated Virus Serotype 8-Mediated Genetic Labeling of Cholangiocytes in the Neonatal Murine Liver.Pharmaceutics. 2020 Apr 13;12(4):351. doi: 10.3390/pharmaceutics12040351. Pharmaceutics. 2020. PMID: 32295003 Free PMC article.
-
Global Analysis of Plasma Lipids Identifies Liver-Derived Acylcarnitines as a Fuel Source for Brown Fat Thermogenesis.Cell Metab. 2017 Sep 5;26(3):509-522.e6. doi: 10.1016/j.cmet.2017.08.006. Cell Metab. 2017. PMID: 28877455 Free PMC article.
-
Fate tracing of hepatocytes in mouse liver.Sci Rep. 2017 Nov 23;7(1):16108. doi: 10.1038/s41598-017-15973-7. Sci Rep. 2017. PMID: 29170436 Free PMC article.
-
Non-Parenchymal Cells and the Extracellular Matrix in Hepatocellular Carcinoma in Non-Alcoholic Fatty Liver Disease.Cancers (Basel). 2023 Feb 18;15(4):1308. doi: 10.3390/cancers15041308. Cancers (Basel). 2023. PMID: 36831649 Free PMC article. Review.
-
Non-Genomic Control of Dynamic MYCN Gene Expression in Liver Cancer.Front Oncol. 2021 Apr 16;10:618515. doi: 10.3389/fonc.2020.618515. eCollection 2020. Front Oncol. 2021. PMID: 33937011 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

