Orphan nuclear receptor oestrogen-related receptor γ (ERRγ) plays a key role in hepatic cannabinoid receptor type 1-mediated induction of CYP7A1 gene expression
- PMID: 26348907
- PMCID: PMC5333639
- DOI: 10.1042/BJ20141494
Orphan nuclear receptor oestrogen-related receptor γ (ERRγ) plays a key role in hepatic cannabinoid receptor type 1-mediated induction of CYP7A1 gene expression
Abstract
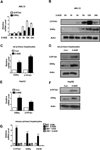
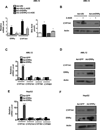
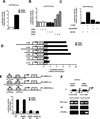
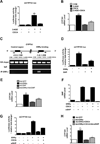
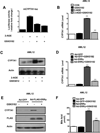
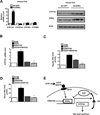
Bile acids are primarily synthesized from cholesterol in the liver and have important roles in dietary lipid absorption and cholesterol homoeostasis. Detailed roles of the orphan nuclear receptors regulating cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid synthesis, have not yet been fully elucidated. In the present study, we report that oestrogen-related receptor γ (ERRγ) is a novel transcriptional regulator of CYP7A1 expression. Activation of cannabinoid receptor type 1 (CB1 receptor) signalling induced ERRγ-mediated transcription of the CYP7A1 gene. Overexpression of ERRγ increased CYP7A1 expression in vitro and in vivo, whereas knockdown of ERRγ attenuated CYP7A1 expression. Deletion analysis of the CYP7A1 gene promoter and a ChIP assay revealed an ERRγ-binding site on the CYP7A1 gene promoter. Small heterodimer partner (SHP) inhibited the transcriptional activity of ERRγ and thus regulated CYP7A1 expression. Overexpression of ERRγ led to increased bile acid levels, whereas an inverse agonist of ERRγ, GSK5182, reduced CYP7A1 expression and bile acid synthesis. Finally, GSK5182 significantly reduced hepatic CB1 receptor-mediated induction of CYP7A1 expression and bile acid synthesis in alcohol-treated mice. These results provide the molecular mechanism linking ERRγ and bile acid metabolism.
Keywords: GSK5182; bile acid; cannabinoid receptors; cholesterol 7α-hydroxylase (CYP7A1); oestrogen-related receptor γ (ERRγ); orphan nuclear receptor; small heterodimer partner (SHP).
© 2015 Authors; published by Portland Press Limited.
Figures
Similar articles
-
An inverse agonist of estrogen-related receptor γ regulates 2-arachidonoylglycerol synthesis by modulating diacylglycerol lipase expression in alcohol-intoxicated mice.Arch Toxicol. 2020 Feb;94(2):427-438. doi: 10.1007/s00204-019-02648-7. Epub 2020 Jan 7. Arch Toxicol. 2020. PMID: 31912162 Free PMC article.
-
Inverse agonist of ERRγ reduces cannabinoid receptor type 1-mediated induction of fibrinogen synthesis in mice with a high-fat diet-intoxicated liver.Arch Toxicol. 2018 Sep;92(9):2885-2896. doi: 10.1007/s00204-018-2270-4. Epub 2018 Jul 17. Arch Toxicol. 2018. PMID: 30019168
-
The Orphan Nuclear Receptor ERRγ Regulates Hepatic CB1 Receptor-Mediated Fibroblast Growth Factor 21 Gene Expression.PLoS One. 2016 Jul 25;11(7):e0159425. doi: 10.1371/journal.pone.0159425. eCollection 2016. PLoS One. 2016. PMID: 27455076 Free PMC article.
-
Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP.J Lipid Res. 2002 Apr;43(4):533-43. J Lipid Res. 2002. PMID: 11907135 Review.
-
Xol INXS: role of the liver X and the farnesol X receptors.Curr Opin Lipidol. 2001 Apr;12(2):113-20. doi: 10.1097/00041433-200104000-00004. Curr Opin Lipidol. 2001. PMID: 11264982 Review.
Cited by
-
An inverse agonist of estrogen-related receptor γ regulates 2-arachidonoylglycerol synthesis by modulating diacylglycerol lipase expression in alcohol-intoxicated mice.Arch Toxicol. 2020 Feb;94(2):427-438. doi: 10.1007/s00204-019-02648-7. Epub 2020 Jan 7. Arch Toxicol. 2020. PMID: 31912162 Free PMC article.
-
ERRγ suppression by Sirt6 alleviates cholestatic liver injury and fibrosis.JCI Insight. 2020 Sep 3;5(17):e137566. doi: 10.1172/jci.insight.137566. JCI Insight. 2020. PMID: 32701506 Free PMC article.
-
An Inverse Agonist GSK5182 Increases Protein Stability of the Orphan Nuclear Receptor ERRγ via Inhibition of Ubiquitination.Int J Mol Sci. 2022 Dec 21;24(1):96. doi: 10.3390/ijms24010096. Int J Mol Sci. 2022. PMID: 36613556 Free PMC article.
-
Hypolipidemic effect of Alisma orientale (Sam.) Juzep on gut microecology and liver transcriptome in diabetic rats.PLoS One. 2020 Oct 9;15(10):e0240616. doi: 10.1371/journal.pone.0240616. eCollection 2020. PLoS One. 2020. PMID: 33035272 Free PMC article.
-
Repression of hepatocyte nuclear factor 4 alpha by AP-1 underlies dyslipidemia associated with retinoic acid.J Lipid Res. 2019 Apr;60(4):794-804. doi: 10.1194/jlr.M088880. Epub 2019 Feb 1. J Lipid Res. 2019. PMID: 30709899 Free PMC article.
References
-
- Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. - PubMed
-
- Yang X, Downes M, Ruth TY, Bookout AL, He W, Straume M, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. - PubMed
-
- Razzaque MA, Masuda N, Maeda Y, Endo Y, Tsukamoto T, Osumi T. Estrogen receptor-related receptor γ has an exceptionally broad specificity of DNA sequence recognition. Gene. 2004;340:275–282. - PubMed
-
- Busch BB, Stevens WC, Martin R, Ordentlich P, Zhou S, Sapp DW, Horlick RA, Mohan R. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor α. J. Med. Chem. 2004;47:5593–5596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

