Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling
- PMID: 26350214
- PMCID: PMC4666394
- DOI: 10.1093/nar/gkv896
Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling
Abstract
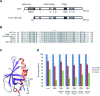
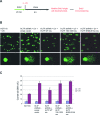
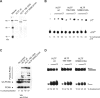
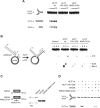
Defects in the ability to respond properly to an unrepaired DNA lesion blocking replication promote genomic instability and cancer. Human HLTF, implicated in error-free replication of damaged DNA and tumour suppression, exhibits a HIRAN domain, a RING domain, and a SWI/SNF domain facilitating DNA-binding, PCNA-polyubiquitin-ligase, and dsDNA-translocase activities, respectively. Here, we investigate the mechanism of HLTF action with emphasis on its HIRAN domain. We found that in cells HLTF promotes the filling-in of gaps left opposite damaged DNA during replication, and this postreplication repair function depends on its HIRAN domain. Our biochemical assays show that HIRAN domain mutant HLTF proteins retain their ubiquitin ligase, ATPase and dsDNA translocase activities but are impaired in binding to a model replication fork. These data and our structural study indicate that the HIRAN domain recruits HLTF to a stalled replication fork, and it also provides the direction for the movement of the dsDNA translocase motor domain for fork reversal. In more general terms, we suggest functional similarities between the HIRAN, the OB, the HARP2, and other domains found in certain motor proteins, which may explain why only a subset of DNA translocases can carry out fork reversal.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

