Excessive Cytolytic Responses Predict Tuberculosis Relapse After Apparently Successful Treatment
- PMID: 26351358
- PMCID: PMC4704670
- DOI: 10.1093/infdis/jiv447
Excessive Cytolytic Responses Predict Tuberculosis Relapse After Apparently Successful Treatment
Abstract
Background: Currently, there are no tools to accurately predict tuberculosis relapse. This study aimed to determine whether patients who experience tuberculosis relapse have different immune responses to mycobacteria in vitro than patients who remain cured for 2 years.
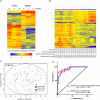
Methods: Patients with an initial episode of pulmonary tuberculosis were recruited in South Africa. Diluted blood, collected at diagnosis and after 2 and 4 weeks of treatment, was cultured with live Mycobacterium tuberculosis for 6 days, and cellular RNA was frozen. Gene expression in samples from 10 patients who subsequently experienced relapse, confirmed by strain genotyping, was compared to that in samples from patients who remained cured, using microarrays.
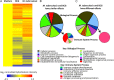
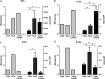
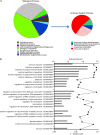
Results: At diagnosis, expression of 668 genes was significantly different in samples from patients who experienced relapse, compared with expression in patients who remained successfully cured; these differences persisted for at least 4 weeks. Gene ontology and biological pathways analyses revealed significant upregulation of genes involved in cytotoxic cell-mediated killing. Results were confirmed by real-time quantitative reverse-transcription polymerase chain reaction analysis in a wider patient cohort.
Conclusions: These data show that patients who will subsequently experience relapse exhibit altered immune responses, including excessively robust cytolytic responses to M. tuberculosis in vitro, at the time of diagnosis, compared with patients who will achieve durable cure. Together with microbiological and clinical indices, these differences could be exploited in drug development.
Keywords: blood; drug development; microarray; patient; transcriptomics.
© The Author 2015. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures
References
-
- Jindani A, Nunn AJ, Enarson DA. Two 8-month regimens of chemotherapy for treatment of newly diagnosed pulmonary tuberculosis: international multicentre randomised trial. Lancet 2004; 364:1244–51. - PubMed
-
- Marx FM, Dunbar R, Enarson DA et al. . The temporal dynamics of relapse and reinfection tuberculosis after successful treatment: a retrospective cohort study. Clin Infect Dis 2014; 58:1676–83. - PubMed
-
- Wallis RS, Wang C, Doherty TM et al. . Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis 2010; 10:68–9. - PubMed
-
- Hesseling AC, Walzl G, Enarson DA et al. . Baseline sputum time to detection predicts month two culture conversion and relapse in non-HIV-infected patients. Int J Tuberc Lung Dis 2010; 14:560–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

