Transcription factors regulating B cell fate in the germinal centre
- PMID: 26352785
- PMCID: PMC4687514
- DOI: 10.1111/cei.12702
Transcription factors regulating B cell fate in the germinal centre
Abstract
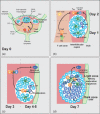
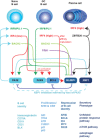
Diversification of the antibody repertoire is essential for the normal operation of the vertebrate adaptive immune system. Following antigen encounter, B cells are activated, proliferate rapidly and undergo two diversification events; somatic hypermutation (followed by selection), which enhances the affinity of the antibody for its cognate antigen, and class-switch recombination, which alters the effector functions of the antibody to adapt the response to the challenge faced. B cells must then differentiate into antibody-secreting plasma cells or long-lived memory B cells. These activities take place in specialized immunological environments called germinal centres, usually located in the secondary lymphoid organs. To complete the germinal centre activities successfully, a B cell adopts a transcriptional programme that allows it to migrate to specific sites within the germinal centre, proliferate, modify its DNA recombination and repair pathways, alter its apoptotic potential and finally undergo terminal differentiation. To co-ordinate these processes, B cells employ a number of 'master regulator' transcription factors which mediate wholesale transcriptomic changes. These master transcription factors are mutually antagonistic and form a complex regulatory network to maintain distinct gene expression programs. Within this network, multiple points of positive and negative feedback ensure the expression of the 'master regulators', augmented by a number of 'secondary' factors that reinforce these networks and sense the progress of the immune response. In this review we will discuss the different activities B cells must undertake to mount a successful T cell-dependent immune response and describe how a regulatory network of transcription factors controls these processes.
Keywords: B cell; cell differentiation; gene regulation; transcription factors; transcriptomics.
© 2015 British Society for Immunology.
Figures

References
-
- Nieuwenhuis P, Opstelten D. Functional anatomy of germinal centers. Am J Anat 1984; 170:421–35. - PubMed
-
- Weyand CM, Goronzy JJ. Ectopic germinal center formation in rheumatoid synovitis. Ann NY Acad Sci 2003; 987:140–9. - PubMed
-
- Coker HA, Durham SR, Gould HJ. Local somatic hypermutation and class switch recombination in the nasal mucosa of allergic rhinitis patients. J Immunol 2003; 171:5602–10. - PubMed
-
- Takhar P, Smurthwaite L, Coker HA et al Allergen drives class switching to IgE in the nasal mucosa in allergic rhinitis. J Immunol 2005; 174:5024–32. - PubMed
-
- Peled JU, Kuang FL, Iglesias‐Ussel MD et al The biochemistry of somatic hypermutation. Annu Rev Immunol 2008; 26:481–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

