Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells
- PMID: 26353929
- PMCID: PMC4694948
- DOI: 10.18632/oncotarget.4925
Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells
Abstract
Background & aims: Liver fibrogenesis - scarring of the liver that can lead to cirrhosis and liver cancer - is characterized by hepatocyte impairment, capillarization of liver sinusoidal endothelial cells (LSECs) and hepatic stellate cell (HSC) activation. To date, the molecular determinants of a healthy human liver cell phenotype remain largely uncharacterized. Here, we assess the transcriptome and the genome-wide promoter methylome specific for purified, non-cultured human hepatocytes, LSECs and HSCs, and investigate the nature of epigenetic changes accompanying transcriptional changes associated with activation of HSCs.
Material and methods: Gene expression profile and promoter methylome of purified, uncultured human liver cells and culture-activated HSCs were respectively determined using Affymetrix HG-U219 genechips and by methylated DNA immunoprecipitation coupled to promoter array hybridization. Histone modification patterns were assessed at the single-gene level by chromatin immunoprecipitation and quantitative PCR.
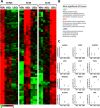
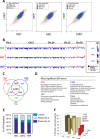
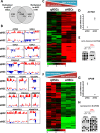
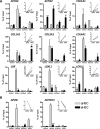
Results: We unveil a DNA-methylation-based epigenetic relationship between hepatocytes, LSECs and HSCs despite their distinct ontogeny. We show that liver cell type-specific DNA methylation targets early developmental and differentiation-associated functions. Integrative analysis of promoter methylome and transcriptome reveals partial concordance between DNA methylation and transcriptional changes associated with human HSC activation. Further, we identify concordant histone methylation and acetylation changes in the promoter and putative novel enhancer elements of genes involved in liver fibrosis.
Conclusions: Our study provides the first epigenetic blueprint of three distinct freshly isolated, human hepatic cell types and of epigenetic changes elicited upon HSC activation.
Keywords: DNA methylation; Pathology Section; epigenetics; hepatic stellate cells; liver fibrosis.
Conflict of interest statement
Authors declare no conflicting interests.
Figures

References
-
- Canbay A, Friedman S, Gores GJ. Apoptosis: The nexus of liver injury and fibrosis. Hepatology. 2004;39:273–278. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

