Rapid deamination of cyclobutane pyrimidine dimer photoproducts at TCG sites in a translationally and rotationally positioned nucleosome in vivo
- PMID: 26354431
- PMCID: PMC4646317
- DOI: 10.1074/jbc.M115.673301
Rapid deamination of cyclobutane pyrimidine dimer photoproducts at TCG sites in a translationally and rotationally positioned nucleosome in vivo
Abstract
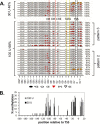
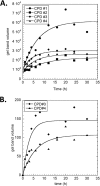
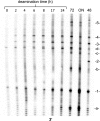
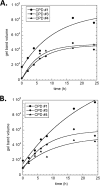
Sunlight-induced C to T mutation hot spots in skin cancers occur primarily at methylated CpG sites that coincide with sites of UV-induced cyclobutane pyrimidine dimer (CPD) formation. The C and 5-methyl-C in CPDs are not stable and deaminate to U and T, respectively, which leads to the insertion of A by the DNA damage bypass polymerase η, thereby defining a probable mechanism for the origin of UV-induced C to T mutations. Deamination rates for T(m)CG CPDs have been found to vary 12-fold with rotational position in a nucleosome in vitro. To determine the influence of nucleosome structure on deamination rates in vivo, we determined the deamination rates of CPDs at TCG sites in a stably positioned nucleosome within the FOS promoter in HeLa cells. A procedure for in vivo hydroxyl radical footprinting with Fe-EDTA was developed, and, together with results from a cytosine methylation protection assay, we determined the translational and rotational positions of the TCG sites. Consistent with the in vitro observations, deamination was slower for one CPD located at an intermediate rotational position compared with two other sites located at outside positions, and all were much faster than for CPDs at non-TCG sites. Photoproduct formation was also highly suppressed at one site, possibly due to its interaction with a histone tail. Thus, it was shown that CPDs of TCG sites deaminate the fastest in vivo and that nucleosomes can modulate both their formation and deamination, which could contribute to the UV mutation hot spots and cold spots.
Keywords: 5-methylcytosine; DNA damage; chromatin structure; histone; mutagenesis mechanism; nucleosome; photobiology.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Pfeifer G. P., You Y. H., and Besaratinia A. (2005) Mutations induced by ultraviolet light. Mutat. Res. 571, 19–31 - PubMed
-
- Vu B., Cannistraro V. J., Sun L., and Taylor J. S. (2006) DNA synthesis past a 5-methylC-containing cis-syn-cyclobutane pyrimidine dimer by yeast pol η is highly nonmutagenic. Biochemistry 45, 9327–9335 - PubMed
-
- Ikehata H., and Ono T. (2011) The mechanisms of UV mutagenesis. J. Radiat. Res. 52, 115–125 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

