Using protein motion to read, write, and erase ubiquitin signals
- PMID: 26354440
- PMCID: PMC4646302
- DOI: 10.1074/jbc.R115.653675
Using protein motion to read, write, and erase ubiquitin signals
Abstract
Eukaryotes use a tiny protein called ubiquitin to send a variety of signals, most often by post-translationally attaching ubiquitins to substrate proteins and to each other, thereby forming polyubiquitin chains. A combination of biophysical, biochemical, and biological studies has shown that complex macromolecular dynamics are central to many aspects of ubiquitin signaling. This review focuses on how equilibrium fluctuations and coordinated motions of ubiquitin itself, the ubiquitin conjugation machinery, and deubiquitinating enzymes enable activity and regulation on many levels, with implications for how such a tiny protein can send so many signals.
Keywords: biophysics; deubiquitylation (deubiquitination); structural biology; ubiquitin; ubiquitylation (ubiquitination).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
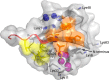
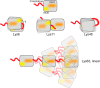
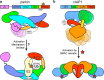
References
-
- Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 - PubMed
-
- Popovic D., Vucic D., and Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 - PubMed
-
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., and Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

