Effects of non-CpG site methylation on DNA thermal stability: a fluorescence study
- PMID: 26354864
- PMCID: PMC4678853
- DOI: 10.1093/nar/gkv884
Effects of non-CpG site methylation on DNA thermal stability: a fluorescence study
Abstract
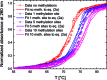
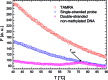
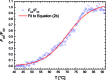
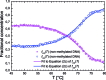
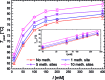
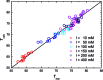
Cytosine methylation is a widespread epigenetic regulation mechanism. In healthy mature cells, methylation occurs at CpG dinucleotides within promoters, where it primarily silences gene expression by modifying the binding affinity of transcription factors to the promoters. Conversely, a recent study showed that in stem cells and cancer cell precursors, methylation also occurs at non-CpG pairs and involves introns and even gene bodies. The epigenetic role of such methylations and the molecular mechanisms by which they induce gene regulation remain elusive. The topology of both physiological and aberrant non-CpG methylation patterns still has to be detailed and could be revealed by using the differential stability of the duplexes formed between site-specific oligonucleotide probes and the corresponding methylated regions of genomic DNA. Here, we present a systematic study of the thermal stability of a DNA oligonucleotide sequence as a function of the number and position of non-CpG methylation sites. The melting temperatures were determined by monitoring the fluorescence of donor-acceptor dual-labelled oligonucleotides at various temperatures. An empirical model that estimates the methylation-induced variations in the standard values of hybridization entropy and enthalpy was developed.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Holliday R., Pugh J.E. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. - PubMed
-
- Riggs A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975;14:9–25. - PubMed
-
- Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. - PubMed
-
- Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

