Fine-scale mapping of the 4q24 locus identifies two independent loci associated with breast cancer risk
- PMID: 26354892
- PMCID: PMC4633342
- DOI: 10.1158/1055-9965.EPI-15-0363
Fine-scale mapping of the 4q24 locus identifies two independent loci associated with breast cancer risk
Abstract
Background: A recent association study identified a common variant (rs9790517) at 4q24 to be associated with breast cancer risk. Independent association signals and potential functional variants in this locus have not been explored.
Methods: We conducted a fine-mapping analysis in 55,540 breast cancer cases and 51,168 controls from the Breast Cancer Association Consortium.
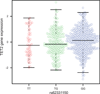
Results: Conditional analyses identified two independent association signals among women of European ancestry, represented by rs9790517 [conditional P = 2.51 × 10(-4); OR, 1.04; 95% confidence interval (CI), 1.02-1.07] and rs77928427 (P = 1.86 × 10(-4); OR, 1.04; 95% CI, 1.02-1.07). Functional annotation using data from the Encyclopedia of DNA Elements (ENCODE) project revealed two putative functional variants, rs62331150 and rs73838678 in linkage disequilibrium (LD) with rs9790517 (r(2) ≥ 0.90) residing in the active promoter or enhancer, respectively, of the nearest gene, TET2. Both variants are located in DNase I hypersensitivity and transcription factor-binding sites. Using data from both The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), we showed that rs62331150 was associated with level of expression of TET2 in breast normal and tumor tissue.
Conclusion: Our study identified two independent association signals at 4q24 in relation to breast cancer risk and suggested that observed association in this locus may be mediated through the regulation of TET2.
Impact: Fine-mapping study with large sample size warranted for identification of independent loci for breast cancer risk.
©2015 American Association for Cancer Research.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


References
-
- iCOGs [Internet] Cambridge (UK): Center for Cancer Genetic Epidemiology, Department of Public Health and Primary Care/Department of Oncology, University of Cambridge; Available from: http://ccge.medschl.cam.ac.uk/research/consortia/icogs/
-
- Tefferi A, Lim KH, Levine R. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;361:1117. author reply -8. - PubMed
-
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360:2289–2301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA54281/CA/NCI NIH HHS/United States
- CA116201/CA/NCI NIH HHS/United States
- CA128978/CA/NCI NIH HHS/United States
- C1287/A10710/CRUK_/Cancer Research UK/United Kingdom
- R01CA148667/CA/NCI NIH HHS/United States
- 16459/CRUK_/Cancer Research UK/United Kingdom
- 1U19 CA148065/CA/NCI NIH HHS/United States
- R01 CA128978/CA/NCI NIH HHS/United States
- R01 CA064277/CA/NCI NIH HHS/United States
- CA098758/CA/NCI NIH HHS/United States
- R01CA64277/CA/NCI NIH HHS/United States
- UM1 CA164920/CA/NCI NIH HHS/United States
- P30 CA68485/CA/NCI NIH HHS/United States
- R37 CA070867/CA/NCI NIH HHS/United States
- 16565/CRUK_/Cancer Research UK/United Kingdom
- R01 CA092447/CA/NCI NIH HHS/United States
- CA122340/CA/NCI NIH HHS/United States
- 1U19 CA148537/CA/NCI NIH HHS/United States
- R01 CA100374/CA/NCI NIH HHS/United States
- CA63464/CA/NCI NIH HHS/United States
- C490/A10124/CRUK_/Cancer Research UK/United Kingdom
- U19 CA148537/CA/NCI NIH HHS/United States
- R01 CA148667/CA/NCI NIH HHS/United States
- R37CA70867/CA/NCI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- C1287/A12014/CRUK_/Cancer Research UK/United Kingdom
- R01 CA063464/CA/NCI NIH HHS/United States
- 16563/CRUK_/Cancer Research UK/United Kingdom
- U01 CA063464/CA/NCI NIH HHS/United States
- R01 CA077398/CA/NCI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- R01 CA132839/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 CA77398/CA/NCI NIH HHS/United States
- 1U19 CA148112/CA/NCI NIH HHS/United States
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- R37CA070867/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- U01 CA098758/CA/NCI NIH HHS/United States
- N01 CN025403/CA/NCI NIH HHS/United States
- CA132839/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA122340/CA/NCI NIH HHS/United States
- C8197/A16565/CRUK_/Cancer Research UK/United Kingdom
- UM1 CA182910/CA/NCI NIH HHS/United States
- C1287/A10118/CRUK_/Cancer Research UK/United Kingdom
- 001/WHO_/World Health Organization/International
- U01 CA164973/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- 16561/CRUK_/Cancer Research UK/United Kingdom
- 10124/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

