Microbial Speciation
- PMID: 26354896
- PMCID: PMC4588065
- DOI: 10.1101/cshperspect.a018143
Microbial Speciation
Abstract
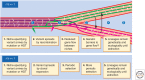
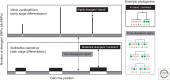
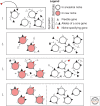
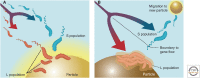
What are species? How do they arise? These questions are not easy to answer and have been particularly controversial in microbiology. Yet, for those microbiologists studying environmental questions or dealing with clinical issues, the ability to name and recognize species, widely considered the fundamental units of ecology, can be practically useful. On a more fundamental level, the speciation problem, the focus here, is more mechanistic and conceptual. What is the origin of microbial species, and what evolutionary and ecological mechanisms keep them separate once they begin to diverge? To what extent are these mechanisms universal across diverse types of microbes, and more broadly across the entire the tree of life? Here, we propose that microbial speciation must be viewed in light of gene flow, which defines units of genetic similarity, and of natural selection, which defines units of phenotype and ecological function. We discuss to what extent ecological and genetic units overlap to form cohesive populations in the wild, based on recent evolutionary modeling and population genomics studies. These studies suggest a continuous "speciation spectrum," which microbial populations traverse in different ways depending on their balance of gene flow and natural selection.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
References
-
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247. - PubMed
-
- Berg OG, Kurland CG. 2002. Evolution of microbial genomes: Sequence acquisition and loss. Mol Biol Evol 19: 2265–2276. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
