Protein Neighbors and Proximity Proteomics
- PMID: 26355100
- PMCID: PMC4638030
- DOI: 10.1074/mcp.R115.052902
Protein Neighbors and Proximity Proteomics
Abstract
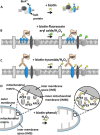
Within cells, proteins can co-assemble into functionally integrated and spatially restricted multicomponent complexes. Often, the affinities between individual proteins are relatively weak, and proteins within such clusters may interact only indirectly with many of their other protein neighbors. This makes proteomic characterization difficult using methods such as immunoprecipitation or cross-linking. Recently, several groups have described the use of enzyme-catalyzed proximity labeling reagents that covalently tag the neighbors of a targeted protein with a small molecule such as fluorescein or biotin. The modified proteins can then be isolated by standard pulldown methods and identified by mass spectrometry. Here we will describe the techniques as well as their similarities and differences. We discuss their applications both to study protein assemblies and to provide a new way for characterizing organelle proteomes. We stress the importance of proteomic quantitation and independent target validation in such experiments. Furthermore, we suggest that there are biophysical and cell-biological principles that dictate the appropriateness of enzyme-catalyzed proximity labeling methods to address particular biological questions of interest.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Ellis R. J. (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11, 114–119 - PubMed
-
- Zimmerman S. B., and Minton A. P. (1993) Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22, 27–65 - PubMed
-
- Han J. D., Bertin N., Hao T., Goldberg D. S., Berriz G. F., Zhang L. V., Dupuy D., Walhout A. J., Cusick M. E., Roth F. P., and Vidal M. (2004) Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430, 88–93 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

