A Robust and Versatile Method of Combinatorial Chemical Synthesis of Gene Libraries via Hierarchical Assembly of Partially Randomized Modules
- PMID: 26355961
- PMCID: PMC4565649
- DOI: 10.1371/journal.pone.0136778
A Robust and Versatile Method of Combinatorial Chemical Synthesis of Gene Libraries via Hierarchical Assembly of Partially Randomized Modules
Abstract
A major challenge in gene library generation is to guarantee a large functional size and diversity that significantly increases the chances of selecting different functional protein variants. The use of trinucleotides mixtures for controlled randomization results in superior library diversity and offers the ability to specify the type and distribution of the amino acids at each position. Here we describe the generation of a high diversity gene library using tHisF of the hyperthermophile Thermotoga maritima as a scaffold. Combining various rational criteria with contingency, we targeted 26 selected codons of the thisF gene sequence for randomization at a controlled level. We have developed a novel method of creating full-length gene libraries by combinatorial assembly of smaller sub-libraries. Full-length libraries of high diversity can easily be assembled on demand from smaller and much less diverse sub-libraries, which circumvent the notoriously troublesome long-term archivation and repeated proliferation of high diversity ensembles of phages or plasmids. We developed a generally applicable software tool for sequence analysis of mutated gene sequences that provides efficient assistance for analysis of library diversity. Finally, practical utility of the library was demonstrated in principle by assessment of the conformational stability of library members and isolating protein variants with HisF activity from it. Our approach integrates a number of features of nucleic acids synthetic chemistry, biochemistry and molecular genetics to a coherent, flexible and robust method of combinatorial gene synthesis.
Conflict of interest statement
Figures



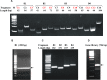

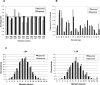


Similar articles
-
Automating gene library synthesis by structure-based combinatorial protein engineering: examples from plant sesquiterpene synthases.Methods Enzymol. 2012;515:21-42. doi: 10.1016/B978-0-12-394290-6.00002-1. Methods Enzymol. 2012. PMID: 22999168
-
Multi-line split DNA synthesis: a novel combinatorial method to make high quality peptide libraries.BMC Biotechnol. 2004 Sep 1;4:19. doi: 10.1186/1472-6750-4-19. BMC Biotechnol. 2004. PMID: 15341664 Free PMC article.
-
Incorporating Synthetic Oligonucleotides via Gene Reassembly (ISOR): a versatile tool for generating targeted libraries.Protein Eng Des Sel. 2007 May;20(5):219-26. doi: 10.1093/protein/gzm014. Epub 2007 May 5. Protein Eng Des Sel. 2007. PMID: 17483523
-
Past and future perspectives of synthetic peptide libraries.Curr Protein Pept Sci. 2008 Oct;9(5):447-67. doi: 10.2174/138920308785915209. Curr Protein Pept Sci. 2008. PMID: 18855697 Review.
-
Steering directed protein evolution: strategies to manage combinatorial complexity of mutant libraries.Environ Microbiol. 2007 Nov;9(11):2645-59. doi: 10.1111/j.1462-2920.2007.01411.x. Environ Microbiol. 2007. PMID: 17922750 Review.
Cited by
-
Preparation of trinucleotide phosphoramidites as synthons for the synthesis of gene libraries.Beilstein J Org Chem. 2018 Feb 13;14:397-406. doi: 10.3762/bjoc.14.28. eCollection 2018. Beilstein J Org Chem. 2018. PMID: 29520304 Free PMC article. Review.
-
Synthesis of fully protected trinucleotide building blocks on a disulphide-linked soluble support.RSC Adv. 2021 Jan 20;11(7):3892-3896. doi: 10.1039/d0ra10941j. eCollection 2021 Jan 19. RSC Adv. 2021. PMID: 35424330 Free PMC article.
-
Split & mix assembly of DNA libraries for ultrahigh throughput on-bead screening of functional proteins.Nucleic Acids Res. 2020 Jun 19;48(11):e63. doi: 10.1093/nar/gkaa270. Nucleic Acids Res. 2020. PMID: 32383757 Free PMC article.
-
Design of an artificial phage-display library based on a new scaffold improved for average stability of the randomized proteins.Sci Rep. 2023 Jan 24;13(1):1339. doi: 10.1038/s41598-023-27710-4. Sci Rep. 2023. PMID: 36693880 Free PMC article.
-
The evolving art of creating genetic diversity: From directed evolution to synthetic biology.Biotechnol Adv. 2021 Sep-Oct;50:107762. doi: 10.1016/j.biotechadv.2021.107762. Epub 2021 May 15. Biotechnol Adv. 2021. PMID: 34000294 Free PMC article. Review.
References
-
- Wong TS, Roccatano D, Schwaneberg U. Steering directed protein evolution: strategies to manage combinatorial complexity of mutant libraries. Environ Microbiol. 2007; 9: 2645–2659. - PubMed
-
- He M, Taussig MJ. Rapid discovery of protein interactions by cell-free protein technologies. Biochem Soc Trans. 2007; 35: 962–965. - PubMed
-
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000; 296: 57–86. - PubMed
-
- Rauchenberger R, Borges E, Thomassen-Wolf E, Rom E, Adar R, Yaniv Y, et al. Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem. 2003; 278: 38194–38205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

