Dynamic Analysis of Gene Expression in Rice Superior and Inferior Grains by RNA-Seq
- PMID: 26355995
- PMCID: PMC4565701
- DOI: 10.1371/journal.pone.0137168
Dynamic Analysis of Gene Expression in Rice Superior and Inferior Grains by RNA-Seq
Abstract
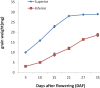
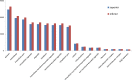
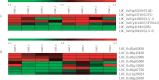
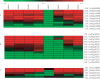
Poor grain filling of inferior grains located on lower secondary panicle branch causes great drop in rice yield and quality. Dynamic gene expression patterns between superior and inferior grains were examined from the view of the whole transcriptome by using RNA-Seq method. In total, 19,442 genes were detected during rice grain development. Genes involved in starch synthesis, grain storage and grain development were interrogated in particular in superior and inferior grains. Of the genes involved in sucrose to starch transformation process, most were expressed at lower level in inferior grains at early filling stage compared to that of superior grains. But at late filling stage, the expression of those genes was higher in inferior grains and lower in superior grains. The same trends were observed in the expression of grain storage protein genes. While, evidence that genes involved in cell cycle showed higher expression in inferior grains during whole period of grain filling indicated that cell proliferation was active till the late filling stage. In conclusion, delayed expression of most starch synthesis genes in inferior grains and low capacity of sink organ might be two important factors causing low filling rate of inferior grain at early filling stage, and shortage of carbohydrate supply was a limiting factor at late filling stage.
Conflict of interest statement
Figures

References
-
- Ishimaru T, Hirose T, Matsuda T, Goto A, Takahashi K, Sasaki H, et al. (2005) Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): comparison of caryopses located at different positions in a panicle. Plant Cell Physiol 46: 620–628. - PubMed
-
- Yang J, Zhang J, Wang Z, Liu K, Wang P (2006) Post-anthesis development of inferior and superior spikelets in rice in relation to abscisic acid and ethylene. J Exp Bot 57: 149–160. - PubMed
-
- Ishimaru T, Matsuda T, Ohsugi R, Yamagishi T (2003) Morphological development of rice caryopses located at the different positions in a panicle from early to middle stage of grain filling. Functional Plant Biology 30: 1139–1149. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

