Dynamic expression of transcription factor Brn3b during mouse cranial nerve development
- PMID: 26356988
- PMCID: PMC4731251
- DOI: 10.1002/cne.23890
Dynamic expression of transcription factor Brn3b during mouse cranial nerve development
Abstract
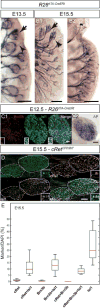
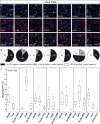
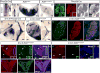
During development, transcription factor combinatorial codes define a large variety of morphologically and physiologically distinct neurons. Such a combinatorial code has been proposed for the differentiation of projection neurons of the somatic and visceral components of cranial nerves. It is possible that individual neuronal cell types are not specified by unique transcription factors but rather emerge through the intersection of their expression domains. Brn3a, Brn3b, and Brn3c, in combination with each other and/or transcription factors of other families, can define subgroups of retinal ganglion cells (RGC), spiral and vestibular ganglia, inner ear and vestibular hair cell neurons in the vestibuloacoustic system, and groups of somatosensory neurons in the dorsal root ganglia. The present study investigates the expression and potential role of the Brn3b transcription factor in cranial nerves and associated nuclei of the brainstem. We report the dynamic expression of Brn3b in the somatosensory component of cranial nerves II, V, VII, and VIII and visceromotor nuclei of nerves VII, IX, and X as well as other brainstem nuclei during different stages of development into adult stage. We find that genetically identified Brn3b(KO) RGC axons show correct but delayed pathfinding during the early stages of embryonic development. However, loss of Brn3b does not affect the anatomy of the other cranial nerves normally expressing this transcription factor.
Keywords: POU domain Brn3b; cranial nerves; facial nerve; glossopharyngeal nerve; optic nerve; transcription; trigeminal nerve; vagus nerve.
© 2015 Wiley Periodicals, Inc.
Conflict of interest statement
No known conflicts of interest.
Figures











Similar articles
-
Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth.Development. 2002 Jan;129(2):467-77. doi: 10.1242/dev.129.2.467. Development. 2002. PMID: 11807038
-
Genetic interactions between Brn3 transcription factors in retinal ganglion cell type specification.PLoS One. 2013 Oct 8;8(10):e76347. doi: 10.1371/journal.pone.0076347. eCollection 2013. PLoS One. 2013. PMID: 24116103 Free PMC article.
-
Transient Expression of Fez Family Zinc Finger 2 Protein Regulates the Brn3b Gene in Developing Retinal Ganglion Cells.J Biol Chem. 2016 Apr 1;291(14):7661-8. doi: 10.1074/jbc.M115.689448. Epub 2016 Feb 9. J Biol Chem. 2016. PMID: 26861874 Free PMC article.
-
Differential expression of Brn3 transcription factors in intrinsically photosensitive retinal ganglion cells in mouse.J Comp Neurol. 2012 Mar 1;520(4):742-55. doi: 10.1002/cne.22765. J Comp Neurol. 2012. PMID: 21935940
-
Patterning and axon guidance of cranial motor neurons.Nat Rev Neurosci. 2007 Nov;8(11):859-71. doi: 10.1038/nrn2254. Nat Rev Neurosci. 2007. PMID: 17948031 Review.
Cited by
-
Molecular codes for cell type specification in Brn3 retinal ganglion cells.Proc Natl Acad Sci U S A. 2017 May 16;114(20):E3974-E3983. doi: 10.1073/pnas.1618551114. Epub 2017 May 2. Proc Natl Acad Sci U S A. 2017. PMID: 28465430 Free PMC article.
-
A Novel Reporter Mouse Uncovers Endogenous Brn3b Expression.Int J Mol Sci. 2019 Jun 14;20(12):2903. doi: 10.3390/ijms20122903. Int J Mol Sci. 2019. PMID: 31197108 Free PMC article.
-
Retinal ganglion cell defects cause decision shifts in visually evoked defense responses.J Neurophysiol. 2020 Nov 1;124(5):1530-1549. doi: 10.1152/jn.00474.2019. Epub 2020 Sep 30. J Neurophysiol. 2020. PMID: 32997561 Free PMC article.
-
Voltage-sensitive dye recording of glossopharyngeal nerve-related synaptic networks in the embryonic mouse brainstem.IBRO Rep. 2019 May 14;6:176-184. doi: 10.1016/j.ibror.2019.05.004. eCollection 2019 Jun. IBRO Rep. 2019. PMID: 31193501 Free PMC article.
-
Optogenetic Control of the Peripheral Nervous System.Cold Spring Harb Perspect Med. 2019 Dec 2;9(12):a034397. doi: 10.1101/cshperspect.a034397. Cold Spring Harb Perspect Med. 2019. PMID: 30745289 Free PMC article. Review.
References
-
- Altschuler SM, Bao X, Bieger D, Hopkins DA, Miselis RR. Viscerotopic representation of the upper alimentary tract in the rat: sensory ganglia and nuclei of the solitary and spinal trigeminal tracts. Journal of Comparative Neurology. 1989;283(2):248–268. - PubMed
-
- Alvarado-Mallart MR, Batini C, Buisseret-Delmas C, Corvisier J. Trigeminal representations of the masticatory and extraocular proprioceptors as revealed by horseradish peroxidase retrograde transport. Experimental brain research. 1975;23(2):167–179. - PubMed
-
- Auclair F, Valdés N, Marchand R. Rhombomere-specific origin of branchial and visceral motoneurons of the facial nerve in the rat embryo. Journal of Comparative Neurology. 1996;369(3):451–461. - PubMed
-
- Ayer-Le Lievre, CS Le, Douarin NM. The early development of cranial sensory ganglia and the potentialities of their component cells studied in quail-chick chimeras. Developmental biology. 1982;94(2):291–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

