Cytokinin-induced promotion of root meristem size in the fern Azolla supports a shoot-like origin of euphyllophyte roots
- PMID: 26358624
- PMCID: PMC5049668
- DOI: 10.1111/nph.13630
Cytokinin-induced promotion of root meristem size in the fern Azolla supports a shoot-like origin of euphyllophyte roots
Abstract
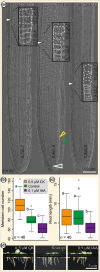
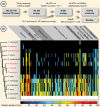
The phytohormones cytokinin and auxin orchestrate the root meristem development in angiosperms by determining embryonic bipolarity. Ferns, having the most basal euphyllophyte root, form neither bipolar embryos nor permanent embryonic primary roots but rather an adventitious root system. This raises the questions of how auxin and cytokinin govern fern root system architecture and whether this can tell us something about the origin of that root. Using Azolla filiculoides, we characterized the influence of IAA and zeatin on adventitious fern root meristems and vasculature by Nomarski microscopy. Simultaneously, RNAseq analyses, yielding 36,091 contigs, were used to uncover how the phytohormones affect root tip gene expression. We show that auxin restricts Azolla root meristem development, while cytokinin promotes it; it is the opposite effect of what is observed in Arabidopsis. Global gene expression profiling uncovered 145 genes significantly regulated by cytokinin or auxin, including cell wall modulators, cell division regulators and lateral root formation coordinators. Our data illuminate both evolution and development of fern roots. Promotion of meristem size through cytokinin supports the idea that root meristems of euphyllophytes evolved from shoot meristems. The foundation of these roots was laid in a postembryonically branching shoot system.
Keywords: RNAseq; apical development; auxin; cytokinin; fern; meristem regulation; root development; root evolution.
© 2015 The Authors. New Phytologist © 2015 New Phytologist Trust.
Figures





References
-
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh Y‐S, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. - PubMed
-
- Amborella Genome Project . 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. - PubMed
-
- Banks JA. 1999. Gametophyte development in ferns. Annual Review of Plant Physiology and Plant Molecular Biology 50: 163–186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

