Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone
- PMID: 26359402
- PMCID: PMC6861171
- DOI: 10.1126/science.aac7202
Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone
Abstract
Podophyllotoxin is the natural product precursor of the chemotherapeutic etoposide, yet only part of its biosynthetic pathway is known. We used transcriptome mining in Podophyllum hexandrum (mayapple) to identify biosynthetic genes in the podophyllotoxin pathway. We selected 29 candidate genes to combinatorially express in Nicotiana benthamiana (tobacco) and identified six pathway enzymes, including an oxoglutarate-dependent dioxygenase that closes the core cyclohexane ring of the aryltetralin scaffold. By coexpressing 10 genes in tobacco-these 6 plus 4 previously discovered-we reconstitute the pathway to (-)-4'-desmethylepipodophyllotoxin (the etoposide aglycone), a naturally occurring lignan that is the immediate precursor of etoposide and, unlike podophyllotoxin, a potent topoisomerase inhibitor. Our results enable production of the etoposide aglycone in tobacco and circumvent the need for cultivation of mayapple and semisynthetic epimerization and demethylation of podophyllotoxin.
Copyright © 2015, American Association for the Advancement of Science.
Figures
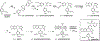
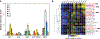

Comment in
-
PLANT BIOCHEMISTRY. Fighting cancer while saving the mayapple.Science. 2015 Sep 11;349(6253):1167-8. doi: 10.1126/science.aad1801. Science. 2015. PMID: 26359390 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- SRA/SRP061783
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

