Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE
- PMID: 26359492
- PMCID: PMC4646338
- DOI: 10.1074/jbc.M115.683334
Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE
Abstract
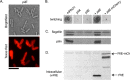
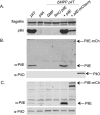
Many bacterial pathogens, including Pseudomonas aeruginosa, use type IVa pili (T4aP) for attachment and twitching motility. T4aP are composed primarily of major pilin subunits, which are repeatedly assembled and disassembled to mediate function. A group of pilin-like proteins, the minor pilins FimU and PilVWXE, prime pilus assembly and are incorporated into the pilus. We showed previously that minor pilin PilE depends on the putative priming subcomplex PilVWX and the non-pilin protein PilY1 for incorporation into pili, and that with FimU, PilE may couple the priming subcomplex to the major pilin PilA, allowing for efficient pilus assembly. Here we provide further support for this model, showing interaction of PilE with other minor pilins and the major pilin. A 1.25 Å crystal structure of PilEΔ1-28 shows a typical type IV pilin fold, demonstrating how it may be incorporated into the pilus. Despite limited sequence identity, PilE is structurally similar to Neisseria meningitidis minor pilins PilXNm and PilVNm, recently suggested via characterization of mCherry fusions to modulate pilus assembly from within the periplasm. A P. aeruginosa PilE-mCherry fusion failed to complement twitching motility or piliation of a pilE mutant. However, in a retraction-deficient strain where surface piliation depends solely on PilE, the fusion construct restored some surface piliation. PilE-mCherry was present in sheared surface fractions, suggesting that it was incorporated into pili. Together, these data provide evidence that PilE, the sole P. aeruginosa equivalent of PilXNm and PilVNm, likely connects a priming subcomplex to the major pilin, promoting efficient assembly of T4aP.
Keywords: Neisseria; Neisseria meningitidis; PilC1; PilY1; Pseudomonas aeruginosa (P. aeruginosa); X-ray crystallography; bacterial adhesion; bacterial pathogenesis; bacterial two-hybrid; minor pilins; pilus assembly; protein-protein interaction; twitching motility; type IV pili.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


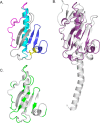

References
-
- Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 - PubMed
-
- Albers S. V., and Pohlschröder M. (2009) Diversity of archaeal type IV pilin-like structures. Extremophiles 13, 403–410 - PubMed
-
- Craig L., Pique M. E., and Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 - PubMed
-
- Ayers M., Howell P. L., and Burrows L. L. (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5, 1203–1218 - PubMed
-
- Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

