Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env
- PMID: 26359989
- PMCID: PMC4587768
- DOI: 10.1016/j.cell.2015.08.035
Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env
Abstract
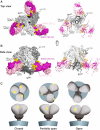
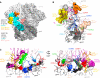
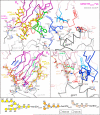
The HIV-1 envelope (Env) spike contains limited epitopes for broadly neutralizing antibodies (bNAbs); thus, most neutralizing antibodies are strain specific. The 8ANC195 epitope, defined by crystal and electron microscopy (EM) structures of bNAb 8ANC195 complexed with monomeric gp120 and trimeric Env, respectively, spans the gp120 and gp41 Env subunits. To investigate 8ANC195's gp41 epitope at higher resolution, we solved a 3.58 Å crystal structure of 8ANC195 complexed with fully glycosylated Env trimer, revealing 8ANC195 insertion into a glycan shield gap to contact gp120 and gp41 glycans and protein residues. To determine whether 8ANC195 recognizes the CD4-bound open Env conformation that leads to co-receptor binding and fusion, one of several known conformations of virion-associated Env, we solved EM structures of an Env/CD4/CD4-induced antibody/8ANC195 complex. 8ANC195 binding partially closed the CD4-bound trimer, confirming structural plasticity of Env by revealing a previously unseen conformation. 8ANC195's ability to bind different Env conformations suggests advantages for potential therapeutic applications.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Capon DJ, Chamow SM, Mordenti J, Marsters SA, Gregory T, Mitsuya H, Byrn RA, Lucas C, Wurm FM, Groopman JE, et al. Designing CD4 immunoadhesins for AIDS therapy. Nature. 1989;337:525–531. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

