Overexpression and knockout of miR-126 both promote leukemogenesis
- PMID: 26361793
- PMCID: PMC4616234
- DOI: 10.1182/blood-2015-04-639062
Overexpression and knockout of miR-126 both promote leukemogenesis
Abstract
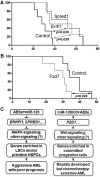
It is generally assumed that gain- and loss-of-function manipulations of a functionally important gene should lead to the opposite phenotypes. We show in this study that both overexpression and knockout of microRNA (miR)-126 surprisingly result in enhanced leukemogenesis in cooperation with the t(8;21) fusion genes AML1-ETO/RUNX1-RUNX1T1 and AML1-ETO9a (a potent oncogenic isoform of AML1-ETO). In accordance with our observation that increased expression of miR-126 is associated with unfavorable survival in patients with t(8;21) acute myeloid leukemia (AML), we show that miR-126 overexpression exhibits a stronger effect on long-term survival and progression of AML1-ETO9a-mediated leukemia stem cells/leukemia initiating cells (LSCs/LICs) in mice than does miR-126 knockout. Furthermore, miR-126 knockout substantially enhances responsiveness of leukemia cells to standard chemotherapy. Mechanistically, miR-126 overexpression activates genes that are highly expressed in LSCs/LICs and/or primitive hematopoietic stem/progenitor cells, likely through targeting ERRFI1 and SPRED1, whereas miR-126 knockout activates genes that are highly expressed in committed, more differentiated hematopoietic progenitor cells, presumably through inducing FZD7 expression. Our data demonstrate that miR-126 plays a critical but 2-faceted role in leukemia and thereby uncover a new layer of miRNA regulation in cancer. Moreover, because miR-126 depletion can sensitize AML cells to standard chemotherapy, our data also suggest that miR-126 represents a promising therapeutic target.
Figures
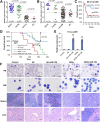
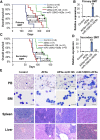
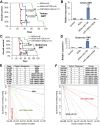
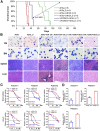


References
-
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

