Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids
- PMID: 26362012
- PMCID: PMC4815224
- DOI: 10.1002/pro.2800
Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids
Abstract
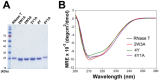
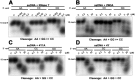
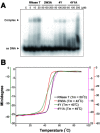
RNase T is a classical member of the DEDDh family of exonucleases with a unique sequence preference in that its 3'-to-5' exonuclease activity is blocked by a 3'-terminal dinucleotide CC in digesting both single-stranded RNA and DNA. Our previous crystal structure analysis of RNase T-DNA complexes show that four phenylalanine residues, F29, F77, F124, and F146, stack with the two 3'-terminal nucleobases. To elucidate if the π-π stacking interactions between aromatic residues and nucleobases play a critical role in sequence-specific protein-nucleic acid recognition, here we mutated two to four of the phenylalanine residues in RNase T to tryptophan (W mutants) and tyrosine (Y mutants). The Escherichia coli strains expressing either the W mutants or the Y mutants had slow growth phenotypes, suggesting that all of these mutants could not fully substitute the function of the wild-type RNase T in vivo. DNA digestion assays revealed W mutants shared similar sequence specificity with wild-type RNase T. However, the Y mutants exhibited altered sequence-dependent activity, digesting ssDNA with both 3'-end CC and GG sequences. Moreover, the W and Y mutants had reduced DNA-binding activity and lower thermal stability as compared to wild-type RNase T. Taken together, our results suggest that the four phenylalanine residues in RNase T not only play critical roles in sequence-specific recognition, but also in overall protein stability. Our results provide the first evidence showing that the π-π stacking interactions between nucleobases and protein aromatic residues may guide the sequence-specific activity for DNA and RNA enzymes.
Keywords: nucleases; protein-DNA interactions; protein-RNA interactions; π-π interactions.
© 2015 The Protein Society.
Figures
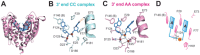

References
-
- Lejeune D, Delsaux N, Charloteaux B, Thomas A, Brasseur R (2005) Protein‐nucleic acid recognition: statistical analysis of atomic interactions and influence of DNA structure. Proteins 61:258–271. - PubMed
-
- Ellis JJ, Broom M, Jones S (2007) Protein‐RNA interactions: structural analysis and functional classes. Proteins 66:903–911. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

