Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response
- PMID: 26362265
- PMCID: PMC4575275
- DOI: 10.1016/j.immuni.2015.06.025
Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response
Abstract
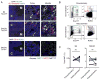
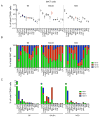
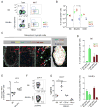
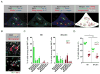
Three subsets of invariant natural killer T (iNKT) cells have been identified, NKT1, NKT2, and NKT17, which produce distinct cytokines when stimulated, but little is known about their localization. Here, we have defined the anatomic localization and systemic distribution of these subsets and measured their cytokine production. Thymic NKT2 cells that produced interleukin-4 (IL-4) at steady state were located in the medulla and conditioned medullary thymocytes. NKT2 cells were abundant in the mesenteric lymph node (LN) of BALB/c mice and produced IL-4 in the T cell zone that conditioned other lymphocytes. Intravenous injection of α-galactosylceramide activated NKT1 cells with vascular access, but not LN or thymic NKT cells, resulting in systemic interferon-γ and IL-4 production, while oral α-galactosylceramide activated NKT2 cells in the mesenteric LN, resulting in local IL-4 release. These findings indicate that the localization of iNKT cells governs their cytokine response both at steady state and upon activation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


References
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annual review of immunology. 2007;25:297–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

