Dense genotyping of immune-related loci in idiopathic inflammatory myopathies confirms HLA alleles as the strongest genetic risk factor and suggests different genetic background for major clinical subgroups
- PMID: 26362759
- PMCID: PMC5300750
- DOI: 10.1136/annrheumdis-2015-208119
Dense genotyping of immune-related loci in idiopathic inflammatory myopathies confirms HLA alleles as the strongest genetic risk factor and suggests different genetic background for major clinical subgroups
Abstract
Objectives: The idiopathic inflammatory myopathies (IIMs) are a heterogeneous group of rare autoimmune diseases characterised by muscle weakness and extramuscular manifestations such as skin rashes and interstitial lung disease. We genotyped 2566 IIM cases of Caucasian descent using the Immunochip; a custom array covering 186 established autoimmune susceptibility loci. The cohort was predominantly comprised of patients with dermatomyositis (DM, n=879), juvenile DM (JDM, n=481), polymyositis (PM, n=931) and inclusion body myositis (n=252) collected from 14 countries through the Myositis Genetics Consortium.
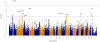
Results: The human leucocyte antigen (HLA) and PTPN22 regions reached genome-wide significance (p<5×10(-8)). Nine regions were associated at a significance level of p<2.25×10(-5), including UBE2L3, CD28 and TRAF6, with evidence of independent effects within STAT4. Analysis of clinical subgroups revealed distinct differences between PM, and DM and JDM. PTPN22 was associated at genome-wide significance with PM, but not DM and JDM, suggesting this effect is driven by PM. Additional suggestive associations including IL18R1 and RGS1 in PM and GSDMB in DM were identified. HLA imputation confirmed that alleles HLA-DRB1*03:01 and HLA-B*08:01 of the 8.1 ancestral haplotype (8.1AH) are most strongly associated with IIM, and provides evidence that amino acids within the HLA, such as HLA-DQB1 position 57 in DM, may explain part of the risk in this locus. Associations with alleles outside the 8.1AH reveal differences between PM, DM and JDM.
Conclusions: This work represents the largest IIM genetic study to date, reveals new insights into the genetic architecture of these rare diseases and suggests different predominating pathophysiology in different clinical subgroups.
Keywords: Dermatomyositis; Gene Polymorphism; Polymyositis.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/
Figures


References
-
- Love LA, Leff RL, Fraser DD, Targoff IN, Dalakas M, Plotz PH, et al. A new approach to the classification of idiopathic inflammatory myopathy: myositis-specific autoantibodies define useful homogeneous patient groups. Medicine (Baltimore) 1991;70:360–74. - PubMed
-
- Chinoy H, Platt H, Lamb JA, Betteridge Z, Gunawardena H, Fertig N, et al. The protein tyrosine phosphatase N22 gene is associated with juvenile and adult idiopathic inflammatory myopathy independent of the HLA 8.1 haplotype in British Caucasian patients. Arthritis Rheum. 2008;58:3247–54. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0900753/MRC_/Medical Research Council/United Kingdom
- MR/K000608/1/MRC_/Medical Research Council/United Kingdom
- 104033/WT_/Wellcome Trust/United Kingdom
- MR/K006312/1/MRC_/Medical Research Council/United Kingdom
- MR/K002279/1/MRC_/Medical Research Council/United Kingdom
- MR/N003322/1/MRC_/Medical Research Council/United Kingdom
- Z01 ES101074/ImNIH/Intramural NIH HHS/United States
- G0600237/MRC_/Medical Research Council/United Kingdom
- P30 CA023108/CA/NCI NIH HHS/United States
- 18474/ARC_/Arthritis Research UK/United Kingdom
- G0100594/MRC_/Medical Research Council/United Kingdom
- G0901461/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

