Genetic variants of the autophagy pathway as prognostic indicators for prostate cancer
- PMID: 26365175
- PMCID: PMC4568463
- DOI: 10.1038/srep14045
Genetic variants of the autophagy pathway as prognostic indicators for prostate cancer
Abstract
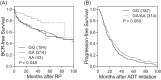
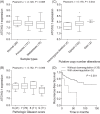
Autophagy is a complex process of autodigestion in conditions of cellular stress, and it might play an important role in the pathophysiology during carcinogenesis. We hypothesize that genetic variants of the autophagy pathway may influence clinical outcomes in prostate cancer patients. We genotyped 40 tagging single-nucleotide polymorphisms (SNPs) from 7 core autophagy pathway genes in 458 localized prostate cancer patients. Multivariate Cox regression was performed to evaluate the independent association of each SNP with disease progression. Positive findings were then replicated in an independent cohort of 504 advanced prostate cancer patients. After adjusting for known clinicopathologic factors, the association between ATG16L1 rs78835907 and recurrence in localized disease [hazard ratio (HR) 0.70, 95% confidence interval (CI) 0.54-0.90, P = 0.006] was replicated in more advanced disease (HR 0.78, 95% CI 0.64-0.95, P = 0.014). Additional integrated in silico analysis suggests that rs78835907 tends to affect ATG16L1 expression, which in turn is correlated with tumor aggressiveness and patient prognosis. In conclusion, genetic variants of the autophagy pathway contribute to the variable outcomes in prostate cancer, and discovery of these novel biomarkers might help stratify patients according to their risk of disease progression.
Figures

References
-
- Draisma G. et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst 95, 868–878 (2003). - PubMed
-
- Simmons M. N., Stephenson A. J. & Klein E. A. Natural history of biochemical recurrence after radical prostatectomy: risk assessment for secondary therapy. Eur Urol 51, 1175–1184 (2007). - PubMed
-
- Akre O. et al. Mortality among men with locally advanced prostate cancer managed with noncurative intent: a nationwide study in PCBaSe Sweden. Eur Urol 60, 554–563 (2011). - PubMed
-
- Walczak J. R. & Carducci M. A. Prostate cancer: a practical approach to current management of recurrent disease. Mayo Clin Proc 82, 243–249 (2007). - PubMed
-
- Ravikumar B. et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90, 1383–1435 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

