TCF1 Is Required for the T Follicular Helper Cell Response to Viral Infection
- PMID: 26365183
- PMCID: PMC4591235
- DOI: 10.1016/j.celrep.2015.08.049
TCF1 Is Required for the T Follicular Helper Cell Response to Viral Infection
Abstract
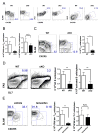
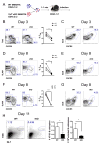
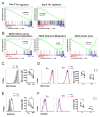
T follicular helper (TFH) and T helper 1 (Th1) cells generated after viral infections are critical for the control of infection and the development of immunological memory. However, the mechanisms that govern the differentiation and maintenance of these two distinct lineages during viral infection remain unclear. We found that viral-specific TFH and Th1 cells showed reciprocal expression of the transcriptions factors TCF1 and Blimp1 early after infection, even before the differential expression of the canonical TFH marker CXCR5. Furthermore, TCF1 was intrinsically required for the TFH cell response to viral infection; in the absence of TCF1, the TFH cell response was severely compromised, and the remaining TCF1-deficient TFH cells failed to maintain TFH-associated transcriptional and metabolic signatures, which were distinct from those in Th1 cells. Mechanistically, TCF1 functioned through forming negative feedback loops with IL-2 and Blimp1. Our findings demonstrate an essential role of TCF1 in TFH cell responses to viral infection.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

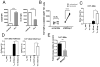

References
-
- Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. Journal of immunology. 2011;187:2089–2092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

