An automated sleep-state classification algorithm for quantifying sleep timing and sleep-dependent dynamics of electroencephalographic and cerebral metabolic parameters
- PMID: 26366107
- PMCID: PMC4562753
- DOI: 10.2147/NSS.S84548
An automated sleep-state classification algorithm for quantifying sleep timing and sleep-dependent dynamics of electroencephalographic and cerebral metabolic parameters
Abstract
Introduction: Rodent sleep research uses electroencephalography (EEG) and electromyography (EMG) to determine the sleep state of an animal at any given time. EEG and EMG signals, typically sampled at >100 Hz, are segmented arbitrarily into epochs of equal duration (usually 2-10 seconds), and each epoch is scored as wake, slow-wave sleep (SWS), or rapid-eye-movement sleep (REMS), on the basis of visual inspection. Automated state scoring can minimize the burden associated with state and thereby facilitate the use of shorter epoch durations.
Methods: We developed a semiautomated state-scoring procedure that uses a combination of principal component analysis and naïve Bayes classification, with the EEG and EMG as inputs. We validated this algorithm against human-scored sleep-state scoring of data from C57BL/6J and BALB/CJ mice. We then applied a general homeostatic model to characterize the state-dependent dynamics of sleep slow-wave activity and cerebral glycolytic flux, measured as lactate concentration.
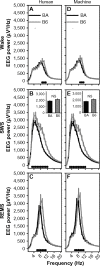
Results: More than 89% of epochs scored as wake or SWS by the human were scored as the same state by the machine, whether scoring in 2-second or 10-second epochs. The majority of epochs scored as REMS by the human were also scored as REMS by the machine. However, of epochs scored as REMS by the human, more than 10% were scored as SWS by the machine and 18 (10-second epochs) to 28% (2-second epochs) were scored as wake. These biases were not strain-specific, as strain differences in sleep-state timing relative to the light/dark cycle, EEG power spectral profiles, and the homeostatic dynamics of both slow waves and lactate were detected equally effectively with the automated method or the manual scoring method. Error associated with mathematical modeling of temporal dynamics of both EEG slow-wave activity and cerebral lactate either did not differ significantly when state scoring was done with automated versus visual scoring, or was reduced with automated state scoring relative to manual classification.
Conclusions: Machine scoring is as effective as human scoring in detecting experimental effects in rodent sleep studies. Automated scoring is an efficient alternative to visual inspection in studies of strain differences in sleep and the temporal dynamics of sleep-related physiological parameters.
Keywords: Bayes classification; EEG; automated scoring; principal component analysis.
Figures









Similar articles
-
The Visual Scoring of Sleep in Infants 0 to 2 Months of Age.J Clin Sleep Med. 2016 Mar;12(3):429-45. doi: 10.5664/jcsm.5600. J Clin Sleep Med. 2016. PMID: 26951412 Free PMC article. Review.
-
The visual scoring of sleep and arousal in infants and children.J Clin Sleep Med. 2007 Mar 15;3(2):201-40. J Clin Sleep Med. 2007. PMID: 17557427 Review.
-
Unsupervised Estimation of Mouse Sleep Scores and Dynamics Using a Graphical Model of Electrophysiological Measurements.Int J Neural Syst. 2016 Jun;26(4):1650017. doi: 10.1142/S0129065716500179. Epub 2016 Feb 16. Int J Neural Syst. 2016. PMID: 27121993
-
The reliability and functional validity of visual and semiautomatic sleep/wake scoring in the Møll-Wistar rat.Sleep. 1994 Mar;17(2):120-31. doi: 10.1093/sleep/17.2.120. Sleep. 1994. PMID: 8036366
-
Multiple classifier systems for automatic sleep scoring in mice.J Neurosci Methods. 2016 May 1;264:33-39. doi: 10.1016/j.jneumeth.2016.02.016. Epub 2016 Feb 27. J Neurosci Methods. 2016. PMID: 26928255 Free PMC article.
Cited by
-
Automatic sleep-wake classification and Parkinson's disease recognition using multifeature fusion with support vector machine.CNS Neurosci Ther. 2024 Apr;30(4):e14708. doi: 10.1111/cns.14708. CNS Neurosci Ther. 2024. PMID: 38600857 Free PMC article.
-
MLS-Net: An Automatic Sleep Stage Classifier Utilizing Multimodal Physiological Signals in Mice.Biosensors (Basel). 2024 Aug 22;14(8):406. doi: 10.3390/bios14080406. Biosensors (Basel). 2024. PMID: 39194635 Free PMC article.
-
SlumberNet: deep learning classification of sleep stages using residual neural networks.Sci Rep. 2024 Feb 27;14(1):4797. doi: 10.1038/s41598-024-54727-0. Sci Rep. 2024. PMID: 38413666 Free PMC article.
-
A novel unsupervised analysis of electrophysiological signals reveals new sleep substages in mice.PLoS Biol. 2018 May 29;16(5):e2003663. doi: 10.1371/journal.pbio.2003663. eCollection 2018 May. PLoS Biol. 2018. PMID: 29813050 Free PMC article.
-
Robust, automated sleep scoring by a compact neural network with distributional shift correction.PLoS One. 2019 Dec 13;14(12):e0224642. doi: 10.1371/journal.pone.0224642. eCollection 2019. PLoS One. 2019. PMID: 31834897 Free PMC article.
References
-
- Stephenson R, Caron AM, Cassel DB, Kostela JC. Automated analysis of sleep-wake state in rats. J Neurosci Methods. 2009;184(2):263–274. - PubMed
-
- Rytkönen KM, Zitting J, Porkka-Heiskanen T. Automated sleep scoring in rats and mice using the naive Bayes classifier. J Neurosci Methods. 2011;202(1):60–64. - PubMed
-
- Brankack J, Kukushka VI, Vyssotski AL, Draguhn A. EEG gamma frequency and sleep-wake scoring in mice: comparing two types of supervised classifiers. Brain Res. 2010;1322:59–71. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

