Triptycene-based small molecules modulate (CAG)·(CTG) repeat junctions
- PMID: 26366282
- PMCID: PMC4538686
- DOI: 10.1039/c5sc01595b
Triptycene-based small molecules modulate (CAG)·(CTG) repeat junctions
Abstract
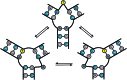
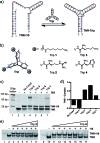
Nucleic acid three-way junctions (3WJs) play key roles in biological processes such as nucleic acid replication in addition to being implicated as dynamic transient intermediates in trinucleotide repeat sequences. Structural modulation of specific nucleic acid junctions could allow for control of biological processes and disease states at the nucleic acid level. Trinucleotide repeat expansions are associated with several neurodegenerative diseases where dynamic slippage is thought to occur during replication, forming transient 3WJ intermediates with the complementary strand. Here, we report triptycene-based molecules that bind to a d(CAG)·(CTG) repeat using a gel shift assay, fluorescence-quenching and circular dichroism.
Figures


References
-
- Howell L. A., Searcey M. Chembiochem. 2009;10:2139–2143. - PubMed
- Sharma A. K., Heemstra J. M. J. Am. Chem. Soc. 2011;133:12426–12429. - PubMed
- Zhang H., Endrizzi J. A., Shu Y., Haque F., Sauter C., Shlyakhtenko L. S., Lyubchenko Y., Guo P., Chi Y. RNAi. 2013;19:1226–1237. - PMC - PubMed
- Teo Y. N., Kool E. T. Chem. Rev. 2012;112:4221–4245. - PMC - PubMed
- Wang F., Lu C. H., Willner I. Chem. Rev. 2014;114:2881–2941. - PubMed
- Stojanovic M. N., de Prada P., Landry D. W. J. Am. Chem. Soc. 2000;122:11547–11548. - PubMed
- Benvin A. L., Creeger Y., Fisher G. W., Ballou B., Waggoner A. S., Armitage B. A. J. Am. Chem. Soc. 2007;129:2025–2034. - PMC - PubMed
- Kellenberger C. A., Wilson S. C., Sales-Lee J., Hammond M. C. J. Am. Chem. Soc. 2013;135:4906–4909. - PMC - PubMed
- Porchetta A., Vallee-Belisle A., Plaxco K. W., Ricci F. J. Am. Chem. Soc. 2012;134:20601–20604. - PMC - PubMed
-
- Leonard C. J., Berns K. I. Prog. Nucleic Acid Res. Mol. Biol. 1994;48:29–52. - PubMed
- Watts J. M., Dang K. K., Gorelick R. J., Leonard C. W., Bess Jr J. W., Swanstrom R., Burch C. L., Weeks K. M. Nature. 2009;460:711–716. - PMC - PubMed
- Bai Y., Tambe A., Zhou K., Doudna J. A. eLife. 2014;3:e03656. - PMC - PubMed
- Guo P. Nat. Nanotechnol. 2010;5:833–842. - PMC - PubMed
- Shu D., Shu Y., Haque F., Abdelmawla S., Guo P. Nat. Nanotechnol. 2011;6:658–667. - PMC - PubMed
- Tian L., Weizmann Y. J. Am. Chem. Soc. 2013;135:1661–1664. - PubMed
- Endo M., Sugiyama H. ChemBioChem. 2009;10:2420–2443. - PubMed
-
- Mirkin S. M. Nature. 2007;447:932–940. - PubMed
- Liu G., Chen X., Bissler J. J., Sinden R. R., Leffak M. Nat. Chem. Biol. 2010;6:652–659. - PMC - PubMed
- Slean M. M., Reddy K., Wu B., Nichol Edamura K., Kekis M., Nelissen F. H., Aspers R. L., Tessari M., Scharer O. D., Wijmenga S. S., Pearson C. E. Biochemistry. 2013;52:773–785. - PMC - PubMed
-
- Chenoweth D. M., Harki D. A., Dervan P. B. Org. Lett. 2009;11:3590–3593. - PMC - PubMed
- Rarig R. A. F., Tran M. N., Chenoweth D. M. J. Am. Chem. Soc. 2013;135:9213–9219. - PubMed
- Dervan P. B. Bioorg. Med. Chem. 2001;9:2215–2235. - PubMed
- Chenoweth D. M., Meier J. L., Dervan P. B. Angew. Chem., Int. Ed. 2013;52:415–418. - PMC - PubMed
- Chenoweth D. M., Dervan P. B. J. Am. Chem. Soc. 2010;132:14521–14529. - PMC - PubMed
- Chenoweth D. M., Dervan P. B. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13175–13179. - PMC - PubMed
- Hirata A., Nokihara K., Kawamoto Y., Bando T., Sasaki A., Ide S., Maeshima K., Kasama T., Sugiyama H. J. Am. Chem. Soc. 2014;136:11546–11554. - PubMed
- Asamitsu S., Kawamoto Y., Hashiya F., Hashiya K., Yamamoto M., Kizaki S., Bando T., Sugiyama H. Bioorg. Med. Chem. 2014;22:4646–4657. - PubMed
- Chenoweth D. M., Poposki J. A., Marques M. A., Dervan P. B. Bioorg. Med. Chem. 2007;15:759–770. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

