Regulatory RNAs and control of epigenetic mechanisms: expectations for cognition and cognitive dysfunction
- PMID: 26366811
- PMCID: PMC5514977
- DOI: 10.2217/epi.15.79
Regulatory RNAs and control of epigenetic mechanisms: expectations for cognition and cognitive dysfunction
Abstract
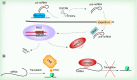
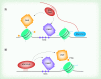
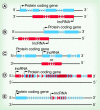
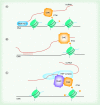
The diverse functions of noncoding RNAs (ncRNAs) can influence virtually every aspect of the transcriptional process including epigenetic regulation of genes. In the CNS, regulatory RNA networks and epigenetic mechanisms have broad relevance to gene transcription changes involved in long-term memory formation and cognition. Thus, it is becoming increasingly clear that multiple classes of ncRNAs impact neuronal development, neuroplasticity, and cognition. Currently, a large gap exists in our knowledge of how ncRNAs facilitate epigenetic processes, and how this phenomenon affects cognitive function. In this review, we discuss recent findings highlighting a provocative role for ncRNAs including lncRNAs and piRNAs in the control of epigenetic mechanisms involved in cognitive function. Furthermore, we discuss the putative roles for these ncRNAs in cognitive disorders such as schizophrenia and Alzheimer's disease.
Keywords: chromatin; epigenetics; long noncoding RNA; neuroplasticity; neuroscience; short noncoding RNA.
Conflict of interest statement
This work was supported by the National Institute of Mental Health (MH082106, MH097909), the UAB Intellectual and Developmental Disabilities Research Center (P30-HD38985) and the Evelyn F McKnight Brain Research Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures
References
-
- Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. - PubMed
-
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat. Rev. Mol. Cell Biol. 2009;10:526–537. - PubMed
-
- Juliandi B, Abematsu M, Nakashima K. Epigenetic regulation in neural stem cell differentiation. Dev. Growth Differ. 2010;52:493–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
