Antibiotic Selection Pressure Determination through Sequence-Based Metagenomics
- PMID: 26369961
- PMCID: PMC4649198
- DOI: 10.1128/AAC.01504-15
Antibiotic Selection Pressure Determination through Sequence-Based Metagenomics
Abstract
The human gut forms a dynamic reservoir of antibiotic resistance genes (ARGs). Treatment with antimicrobial agents has a significant impact on the intestinal resistome and leads to enhanced horizontal transfer and selection of resistance. We have monitored the development of intestinal ARGs over a 6-day course of ciprofloxacin (Cp) treatment in two healthy individuals by using sequenced-based metagenomics and different ARG quantification methods. Fixed- and random-effect models were applied to determine the change in ARG abundance per defined daily dose of Cp as an expression of the respective selection pressure. Among various shifts in the composition of the intestinal resistome, we found in one individual a strong positive selection for class D beta-lactamases which were partly located on a mobile genetic element. Furthermore, a trend to a negative selection has been observed with class A beta-lactamases (-2.66 hits per million sample reads/defined daily dose; P = 0.06). By 4 weeks after the end of treatment, the composition of ARGs returned toward their initial state but to a different degree in both subjects. We present here a novel analysis algorithm for the determination of antibiotic selection pressure which can be applied in clinical settings to compare therapeutic regimens regarding their effect on the intestinal resistome. This information is of critical importance for clinicians to choose antimicrobial agents with a low selective force on their patients' intestinal ARGs, likely resulting in a diminished spread of resistance and a reduced burden of hospital-acquired infections with multidrug-resistant pathogens.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
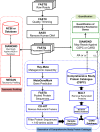
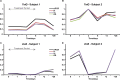
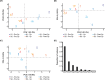

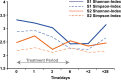
References
-
- European Centre for Disease Prevention and Control. 2014. Point prevalence survey of healthcare-associated infections and antimicrobial use in European long-term care facilities. April–May 2013. European Centre for Disease Prevention and Control, Stockholm, Sweden.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

