Reference-free compression of high throughput sequencing data with a probabilistic de Bruijn graph
- PMID: 26370285
- PMCID: PMC4570262
- DOI: 10.1186/s12859-015-0709-7
Reference-free compression of high throughput sequencing data with a probabilistic de Bruijn graph
Abstract
Background: Data volumes generated by next-generation sequencing (NGS) technologies is now a major concern for both data storage and transmission. This triggered the need for more efficient methods than general purpose compression tools, such as the widely used gzip method.
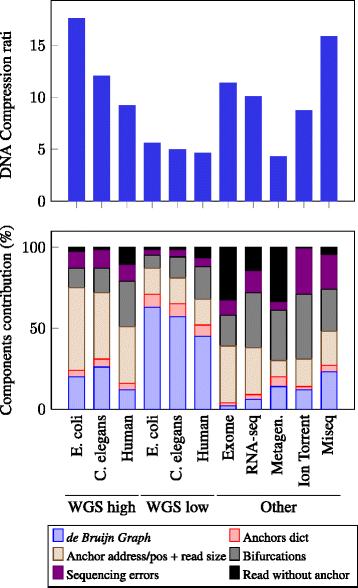
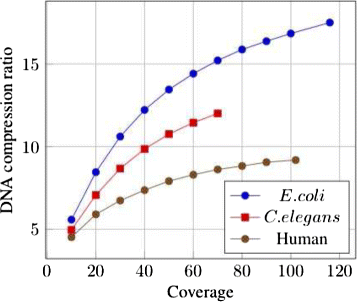
Results: We present a novel reference-free method meant to compress data issued from high throughput sequencing technologies. Our approach, implemented in the software LEON, employs techniques derived from existing assembly principles. The method is based on a reference probabilistic de Bruijn Graph, built de novo from the set of reads and stored in a Bloom filter. Each read is encoded as a path in this graph, by memorizing an anchoring kmer and a list of bifurcations. The same probabilistic de Bruijn Graph is used to perform a lossy transformation of the quality scores, which allows to obtain higher compression rates without losing pertinent information for downstream analyses.
Conclusions: LEON was run on various real sequencing datasets (whole genome, exome, RNA-seq or metagenomics). In all cases, LEON showed higher overall compression ratios than state-of-the-art compression software. On a C. elegans whole genome sequencing dataset, LEON divided the original file size by more than 20. LEON is an open source software, distributed under GNU affero GPL License, available for download at http://gatb.inria.fr/software/leon/.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

