Clinical and molecular epidemiology of human rhinovirus infections in patients with hematologic malignancy
- PMID: 26370315
- PMCID: PMC4750469
- DOI: 10.1016/j.jcv.2015.07.309
Clinical and molecular epidemiology of human rhinovirus infections in patients with hematologic malignancy
Abstract
Background: Human rhinoviruses (HRVs) are common causes of upper respiratory tract infection (URTI) in hematologic malignancy (HM) patients. Predictors of lower respiratory tract infection (LRTI) including the impact of HRV species and types are poorly understood.
Objectives: This study aims to describe the clinical and molecular epidemiology of HRV infections among HM patients.
Study design: From April 2012-March 2013, HRV-positive respiratory specimens from symptomatic HM patients were molecularly characterized by analysis of partial viral protein 1 (VP1) or VP4 gene sequence. HRV LRTI risk-factors and outcomes were analyzed using multivariable logistic regression.
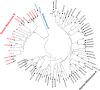
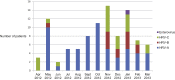
Results: One hundred and ten HM patients presented with HRV URTI (n=78) and HRV LRTI (n=32). Hypoalbuminemia (OR 3.0; 95% CI, 1.0-9.2; p=0.05) was independently associated with LRTI, but other clinical and laboratory markers of host immunity did not differ between patients with URTI versus LRTI. Detection of bacterial co-pathogens was common in LRTI cases (25%). Among 92 typeable respiratory specimens, there were 58 (64%) HRV-As, 12 (13%) HRV-Bs, and 21 (23%) HRV-Cs, and one Enterovirus 68. LRTI rates among HRV-A (29%), HRV-B (17%), and HRV-C (29%) were similar. HRV-A infections occurred year-round while HRV-B and HRV-C infections clustered in the late fall and winter.
Conclusions: HRVs are associated with LRTI in HM patients. Illness severity is not attributable to specific HRV species or types. The frequent detection of bacterial co-pathogens in HRV LRTIs further substantiates the hypothesis that HRVs predispose to bacterial superinfection of the lower airways, similar to that of other community-acquired respiratory viruses.
Keywords: Hematologic malignancy; Human rhinovirus; Immunocompromised hosts; Rhinovirus species; Viral pneumonia.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures

References
-
- Iwane M.K., Prill M.M., Lu X. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young U.S. children. J. Infect. Dis. 2011;204:1702–1710. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

