Outer membrane protein biogenesis in Gram-negative bacteria
- PMID: 26370935
- PMCID: PMC4632599
- DOI: 10.1098/rstb.2015.0023
Outer membrane protein biogenesis in Gram-negative bacteria
Abstract
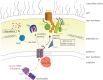
Gram-negative bacteria contain a double membrane which serves for both protection and for providing nutrients for viability. The outermost of these membranes is called the outer membrane (OM), and it contains a host of fully integrated membrane proteins which serve essential functions for the cell, including nutrient uptake, cell adhesion, cell signalling and waste export. For pathogenic strains, many of these outer membrane proteins (OMPs) also serve as virulence factors for nutrient scavenging and evasion of host defence mechanisms. OMPs are unique membrane proteins in that they have a β-barrel fold and can range in size from 8 to 26 strands, yet can still serve many different functions for the cell. Despite their essential roles in cell survival and virulence, the exact mechanism for the biogenesis of these OMPs into the OM has remained largely unknown. However, the past decade has witnessed significant progress towards unravelling the pathways and mechanisms necessary for moulding a nascent polypeptide into a functional OMP within the OM. Here, we will review some of these recent discoveries that have advanced our understanding of the biogenesis of OMPs in Gram-negative bacteria, starting with synthesis in the cytoplasm to folding and insertion into the OM.
Keywords: insertase; lateral gate; outer membrane; protein folding; β-barrel assembly machinery complex; β-barrel membrane protein.
© 2015 The Author(s).
Figures

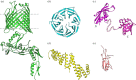
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
