The Proteome Profiles of the Cerebellum of Juvenile, Adult and Aged Rats--An Ontogenetic Study
- PMID: 26370973
- PMCID: PMC4613263
- DOI: 10.3390/ijms160921454
The Proteome Profiles of the Cerebellum of Juvenile, Adult and Aged Rats--An Ontogenetic Study
Abstract
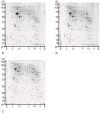
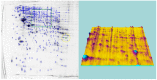
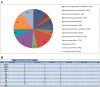
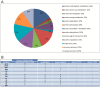
In this study, we searched for proteins that change their expression in the cerebellum (Ce) of rats during ontogenesis. This study focuses on the question of whether specific proteins exist which are differentially expressed with regard to postnatal stages of development. A better characterization of the microenvironment and its development may result from these study findings. A differential two-dimensional polyacrylamide gel electrophoresis (2DE) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis of the samples revealed that the number of proteins of the functional classes differed depending on the developmental stages. Especially members of the functional classes of biosynthesis, regulatory proteins, chaperones and structural proteins show the highest differential expression within the analyzed stages of development. Therefore, members of these functional protein groups seem to be involved in the development and differentiation of the Ce within the analyzed development stages. In this study, changes in the expression of proteins in the Ce at different postnatal developmental stages (postnatal days (P) 7, 90, and 637) could be observed. At the same time, an identification of proteins which are involved in cell migration and differentiation was possible. Especially proteins involved in processes of the biosynthesis and regulation, the dynamic organization of the cytoskeleton as well as chaperones showed a high amount of differentially expressed proteins between the analyzed dates.
Keywords: brain; cerebellum; development; proteomics; rat.
Figures
Similar articles
-
The proteome profiles of the olfactory bulb of juvenile, adult and aged rats - an ontogenetic study.Proteome Sci. 2015 Feb 15;13:8. doi: 10.1186/s12953-014-0058-x. eCollection 2015. Proteome Sci. 2015. PMID: 25709559 Free PMC article.
-
Proteomic analysis reveals differentially secreted proteins in the urine from patients with clear cell renal cell carcinoma.Urol Oncol. 2016 Jan;34(1):5.e11-25. doi: 10.1016/j.urolonc.2015.07.016. Epub 2015 Sep 26. Urol Oncol. 2016. PMID: 26420021
-
Identification of differentially expressed proteins of gamma-ray irradiated rat intestinal epithelial IEC-6 cells by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry.Proteomics. 2005 Feb;5(2):426-32. doi: 10.1002/pmic.200400932. Proteomics. 2005. PMID: 15700242
-
Quantitative differential proteomics of yeast extracellular matrix: there is more to it than meets the eye.BMC Microbiol. 2015 Nov 25;15:271. doi: 10.1186/s12866-015-0550-1. BMC Microbiol. 2015. PMID: 26608260 Free PMC article.
-
Differential proteome of the striatum from hemiparkinsonian rats displays vivid structural remodeling processes.J Proteome Res. 2010 Sep 3;9(9):4671-87. doi: 10.1021/pr100389u. J Proteome Res. 2010. PMID: 20666516
Cited by
-
Comparison of NMDA and AMPA Channel Expression and Function between Embryonic and Adult Neurons Utilizing Microelectrode Array Systems.ACS Biomater Sci Eng. 2017 Dec 11;3(12):3525-3533. doi: 10.1021/acsbiomaterials.7b00596. Epub 2017 Nov 13. ACS Biomater Sci Eng. 2017. PMID: 29250595 Free PMC article.
-
Membrane Protein Identification in Rodent Brain Tissue Samples and Acute Brain Slices.Cells. 2019 May 8;8(5):423. doi: 10.3390/cells8050423. Cells. 2019. PMID: 31072038 Free PMC article.
-
Towards Differential Connectomics with NeuroVIISAS.Neuroinformatics. 2019 Jan;17(1):163-179. doi: 10.1007/s12021-018-9389-6. Neuroinformatics. 2019. PMID: 30014279
References
-
- Ghez C., Fahn S. The cerebellum. In: Kandel E.R., Schwartz J.H., editors. Principles of Neural Science. Elsevier; New York, NY, USA: 1985.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

