The Arabidopsis Chloroplast Stromal N-Terminome: Complexities of Amino-Terminal Protein Maturation and Stability
- PMID: 26371235
- PMCID: PMC4634096
- DOI: 10.1104/pp.15.01214
The Arabidopsis Chloroplast Stromal N-Terminome: Complexities of Amino-Terminal Protein Maturation and Stability
Abstract
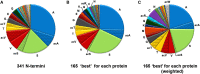
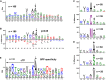
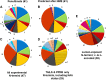
Protein amino (N) termini are prone to modifications and are major determinants of protein stability in bacteria, eukaryotes, and perhaps also in chloroplasts. Most chloroplast proteins undergo N-terminal maturation, but this is poorly understood due to insufficient experimental information. Consequently, N termini of mature chloroplast proteins cannot be accurately predicted. This motivated an extensive characterization of chloroplast protein N termini in Arabidopsis (Arabidopsis thaliana) using terminal amine isotopic labeling of substrates and mass spectrometry, generating nearly 14,000 tandem mass spectrometry spectra matching to protein N termini. Many nucleus-encoded plastid proteins accumulated with two or three different N termini; we evaluated the significance of these different proteoforms. Alanine, valine, threonine (often in N-α-acetylated form), and serine were by far the most observed N-terminal residues, even after normalization for their frequency in the plastid proteome, while other residues were absent or highly underrepresented. Plastid-encoded proteins showed a comparable distribution of N-terminal residues, but with a higher frequency of methionine. Infrequent residues (e.g. isoleucine, arginine, cysteine, proline, aspartate, and glutamate) were observed for several abundant proteins (e.g. heat shock proteins 70 and 90, Rubisco large subunit, and ferredoxin-glutamate synthase), likely reflecting functional regulation through their N termini. In contrast, the thylakoid lumenal proteome showed a wide diversity of N-terminal residues, including those typically associated with instability (aspartate, glutamate, leucine, and phenylalanine). We propose that, after cleavage of the chloroplast transit peptide by stromal processing peptidase, additional processing by unidentified peptidases occurs to avoid unstable or otherwise unfavorable N-terminal residues. The possibility of a chloroplast N-end rule is discussed.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

