The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development
- PMID: 26371508
- PMCID: PMC4566030
- DOI: 10.7554/eLife.08954
The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development
Abstract
Tissue- and cell-type-specific regulators of alternative splicing (AS) are essential components of posttranscriptional gene regulation, necessary for normal cellular function, patterning, and development. Mice with ablation of Epithelial splicing regulatory protein (Esrp1) develop cleft lip and palate. Loss of both Esrp1 and its paralog Esrp2 results in widespread developmental defects with broad implications to human disease. Deletion of the Esrps in the epidermis revealed their requirement for establishing a proper skin barrier, a primary function of epithelial cells comprising the epidermis. We profiled the global Esrp-mediated splicing regulatory program in epidermis, which revealed large-scale programs of epithelial cell-type-specific splicing required for epithelial cell functions. These mice represent a valuable model for evaluating the essential role for AS in development and function of epithelial cells, which play essential roles in tissue homeostasis in numerous organs, and provide a genetic tool to evaluate important functional properties of epithelial-specific splice variants in vivo.
Keywords: alternative splicing; chromosomes; cleft lip associated with cleft palate (CL/P); developmental biology; epithelial barrier defect; epithelial splicing regulatory proteins; genes; mouse; mouse gene knockout; stem cells.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
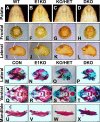

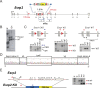




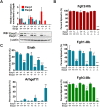





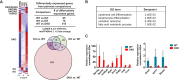

References
-
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, Lyons KM, Mishina Y, Seykora JT, Crenshaw EB, III, Millar SE. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. - DOI - PubMed
-
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. The Journal of Clinical Investigation. 2011;121:1064–1074. doi: 10.1172/JCI44540. - DOI - PMC - PubMed
-
- Carstens RP, Wagner EJ, Garcia-Blanco MA. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Molecular and Cellular Biology. 2000;20:7388–7400. doi: 10.1128/MCB.20.19.7388-7400.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM088342/GM/NIGMS NIH HHS/United States
- R01 GM088809/GM/NIGMS NIH HHS/United States
- R01 GM105431/GM/NIGMS NIH HHS/United States
- F32DK098917/DK/NIDDK NIH HHS/United States
- P30 AR057217/AR/NIAMS NIH HHS/United States
- R01 DE024749/DE/NIDCR NIH HHS/United States
- F32 DK098917/DK/NIDDK NIH HHS/United States
- P30 DK050306/DK/NIDDK NIH HHS/United States
- P30 AR050950/AR/NIAMS NIH HHS/United States
- T32DK700638/DK/NIDDK NIH HHS/United States
- T32 GM008216/GM/NIGMS NIH HHS/United States
- R56 AR066741/AR/NIAMS NIH HHS/United States
- R01 AR066741/AR/NIAMS NIH HHS/United States
- R56 DE024749/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

