Genetic research in autism spectrum disorders
- PMID: 26371945
- PMCID: PMC4650984
- DOI: 10.1097/MOP.0000000000000278
Genetic research in autism spectrum disorders
Abstract
Purpose of review: The recent explosion of genetic findings in autism spectrum disorder (ASD) research has improved knowledge of the disorder's underlying biology and etiologic architecture. This review introduces concepts and results from recent genetic studies and discusses the manner in which those findings can influence the trajectory of ASD research.
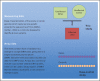
Recent findings: Large consortium studies have associated ASDs with many types of genetic risk factors, including common polygenic risk, de novo single nucleotide variants, copy number variants, and rare inherited variants. In aggregate, these results confirm the heterogeneity and complexity of ASDs. The rare variant findings in particular point to genes and pathways that begin to bridge the gap between behavior and biology.
Summary: Genetic studies have the potential to identify the biological underpinnings of ASDs and other neuropsychiatric disorders. The data they generate are already being used to examine disease pathways and pathogenesis. The results also speak to ASD heterogeneity and, in the future, may be used to stratify research studies and treatment trials.
Figures

References
-
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet 2011; 156:255–274. - PubMed
-
- Gaugler T, Klei L, Sanders SJ, et al. Most genetic risk for autism resides with common variation. Nat Genet 2014; 46:881–885. - PMC - PubMed
-
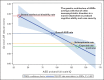
This study reported that most of the genetic influences on ASD risk reflect common, inherited variation. They estimate the proportion of ASD risk attributable to multiple types of genetic variants and find that rare de novo events can have large individual effects but have a limited contribution to risk at a population level.
-
- Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de-novo coding mutations to autism spectrum disorder. Nature 2014; 515:216–221. - PMC - PubMed
-
This study, copublished with De Rubeis et al., uses large-scale whole-exome sequencing technology to examine the contribution of inherited and germline de novo mutations to ASD risk. The authors identified high-confidence gene targets and suggested that de novo missense mutations contribute to a significant number of diagnoses.
-
- De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 515:209–215. - PMC - PubMed
-
This study, copublished with Iossifov et al., uses large-scale whole-exome sequencing technology to examine the contribution of inherited and germline de novo mutations to ASD risk. The authors identified hundreds of de novo LoF mutations, which were estimated to contribute to ASD risk in more than 5% of individuals with autism. The genes harboring de novo LoF mutations were reported to affect synaptic, transcriptional, and chromatin remodeling pathways.
-
- Risch N, Hoffmann TJ, Anderson M, et al. Familial recurrence of autism spectrum disorder: evaluating genetic and environmental contributions. Am J Psychiatry 2014; 171:1206–1213. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

