CDKN3 mRNA as a Biomarker for Survival and Therapeutic Target in Cervical Cancer
- PMID: 26372210
- PMCID: PMC4570808
- DOI: 10.1371/journal.pone.0137397
CDKN3 mRNA as a Biomarker for Survival and Therapeutic Target in Cervical Cancer
Abstract
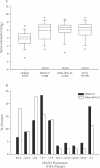
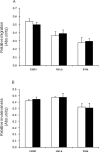
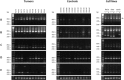
The cyclin-dependent kinase inhibitor 3 (CDKN3) gene, involved in mitosis, is upregulated in cervical cancer (CC). We investigated CDKN3 mRNA as a survival biomarker and potential therapeutic target for CC. CDKN3 mRNA was measured in 134 CC and 25 controls by quantitative PCR. A 5-year survival study was conducted in 121 of these CC patients. Furthermore, CDKN3-specific siRNAs were used to investigate whether CDKN3 is involved in proliferation, migration, and invasion in CC-derived cell lines (SiHa, CaSki, HeLa). CDKN3 mRNA was on average 6.4-fold higher in tumors than in controls (p = 8 x 10-6, Mann-Whitney). A total of 68.2% of CC patients over expressing CDKN3 gene (fold change ≥ 17) died within two years of diagnosis, independent of the clinical stage and HPV type (Hazard Ratio = 5.0, 95% CI: 2.5-10, p = 3.3 x 10-6, Cox proportional-hazards regression). In contrast, only 19.2% of the patients with lower CDKN3 expression died in the same period. In vitro inactivation of CDKN3 decreased cell proliferation on average 67%, although it had no effect on cell migration and invasion. CDKN3 mRNA may be a good survival biomarker and potential therapeutic target in CC.
Conflict of interest statement
Figures



Similar articles
-
Targeting CDKN3 in cervical cancer.Expert Opin Ther Targets. 2014 Oct;18(10):1149-62. doi: 10.1517/14728222.2014.941808. Epub 2014 Aug 25. Expert Opin Ther Targets. 2014. PMID: 25152075 Review.
-
CDKN3 expression is negatively associated with pathological tumor stage and CDKN3 inhibition promotes cell survival in hepatocellular carcinoma.Mol Med Rep. 2016 Aug;14(2):1509-14. doi: 10.3892/mmr.2016.5410. Epub 2016 Jun 17. Mol Med Rep. 2016. PMID: 27314282 Free PMC article.
-
CDKN3 knockdown reduces cell proliferation, invasion and promotes apoptosis in human ovarian cancer.Int J Clin Exp Pathol. 2015 May 1;8(5):4535-44. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26191143 Free PMC article.
-
Integrated transcriptomics explored the cancer-promoting genes CDKN3 in esophageal squamous cell cancer.J Cardiothorac Surg. 2021 May 27;16(1):148. doi: 10.1186/s13019-021-01534-7. J Cardiothorac Surg. 2021. PMID: 34044866 Free PMC article.
-
Expression and alternative splicing of the cyclin-dependent kinase inhibitor-3 gene in human cancer.Int J Biochem Cell Biol. 2017 Oct;91(Pt B):98-101. doi: 10.1016/j.biocel.2017.05.013. Epub 2017 May 11. Int J Biochem Cell Biol. 2017. PMID: 28504190 Free PMC article. Review.
Cited by
-
PSMD12 interacts with CDKN3 and facilitates pancreatic cancer progression.Cancer Gene Ther. 2023 Aug;30(8):1072-1083. doi: 10.1038/s41417-023-00609-y. Epub 2023 Apr 10. Cancer Gene Ther. 2023. PMID: 37037907
-
Development of a novel lipid metabolism-based risk score model in hepatocellular carcinoma patients.BMC Gastroenterol. 2021 Feb 12;21(1):68. doi: 10.1186/s12876-021-01638-3. BMC Gastroenterol. 2021. PMID: 33579192 Free PMC article.
-
Multi-Omics Data Analysis Identifies Prognostic Biomarkers across Cancers.Med Sci (Basel). 2023 Jun 27;11(3):44. doi: 10.3390/medsci11030044. Med Sci (Basel). 2023. PMID: 37489460 Free PMC article.
-
Cyclin-Dependent Kinase Inhibitor 3 Promotes Cancer Cell Proliferation and Tumorigenesis in Nasopharyngeal Carcinoma by Targeting p27.Oncol Res. 2017 Nov 2;25(9):1431-1440. doi: 10.3727/096504017X14835311718295. Epub 2017 Jan 20. Oncol Res. 2017. PMID: 28109073 Free PMC article.
-
CDKN3 promotes cell proliferation, invasion and migration by activating the AKT signaling pathway in esophageal squamous cell carcinoma.Oncol Lett. 2020 Jan;19(1):542-548. doi: 10.3892/ol.2019.11077. Epub 2019 Nov 11. Oncol Lett. 2020. PMID: 31897169 Free PMC article.
References
-
- Bruni L, Barrionuevo-Rosas L, Albero G, Aldea M, Serrano B, Valencia S, et al. ICO Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in the World. Summary Report 2014-02-20. [Data Accessed]
-
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available: http://globocan.iarc.fr, accessed on day/month/year.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

