Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells
- PMID: 26372956
- PMCID: PMC4586827
- DOI: 10.1073/pnas.1512869112
Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells
Abstract
Compensatory growth and regeneration of skeletal muscle is dependent on the resident stem cell population, satellite cells (SCs). Self-renewal and maintenance of the SC niche is coordinated by the paired-box transcription factor Pax7, and yet continued expression of this protein inhibits the myoblast differentiation program. As such, the reduction or removal of Pax7 may denote a key prerequisite for SCs to abandon self-renewal and acquire differentiation competence. Here, we identify caspase 3 cleavage inactivation of Pax7 as a crucial step for terminating the self-renewal process. Inhibition of caspase 3 results in elevated Pax7 protein and SC self-renewal, whereas caspase activation leads to Pax7 cleavage and initiation of the myogenic differentiation program. Moreover, in vivo inhibition of caspase 3 activity leads to a profound disruption in skeletal muscle regeneration with an accumulation of SCs within the niche. We have also noted that casein kinase 2 (CK2)-directed phosphorylation of Pax7 attenuates caspase-directed cleavage. Together, these results demonstrate that SC fate is dependent on opposing posttranslational modifications of the Pax7 protein.
Keywords: Pax7; casein kinase 2; caspase; satellite cells; self-renewal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
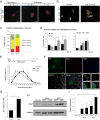
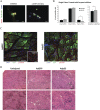
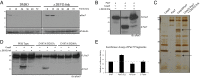

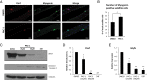
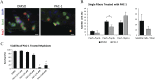
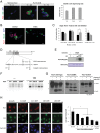

References
-
- Chargé SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84(1):209–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

