Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition
- PMID: 26373314
- PMCID: PMC4609186
- DOI: 10.15252/embj.201591569
Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition
Abstract
Many bacteria use small RNAs (sRNAs) and the RNA chaperone Hfq to regulate mRNA stability and translation. Hfq, a ring-shaped homohexamer, has multiple faces that can bind both sRNAs and their mRNA targets. We find that Hfq has at least two distinct ways in which it interacts with sRNAs; these different binding properties have strong effects on the stability of the sRNA in vivo and the sequence requirements of regulated mRNAs. Class I sRNAs depend on proximal and rim Hfq sites for stability and turn over rapidly. Class II sRNAs are more stable and depend on the proximal and distal Hfq sites for stabilization. Using deletions and chimeras, we find that while Class I sRNAs regulate mRNA targets with previously defined ARN repeats, Class II sRNAs regulate mRNAs carrying UA-rich rim-binding sites. We discuss how these different binding modes may correlate with different roles in the cell, with Class I sRNAs acting as emergency responders and Class II sRNAs acting as silencers.
Keywords: ChiX; Hfq; MgrR; RyhB.
Published 2015. This article is a U.S. Government work and is in the public domain in the USA.
Figures

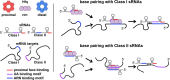

Reported secondary structure of the wild-type ChiX (Rasmussen et al, 2009) and predicted secondary structure of the corresponding ARN deletion mutant (ChiXΔARN). Predicted RNA structures in all figures were determined as described in Reuter and Mathews (2010). Color codes show the ARN motifs in blue, the base-pairing region to the chiP target (Rasmussen et al, 2009) in yellow, and the U-rich region of the rho-independent terminator in red.
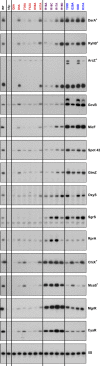
Northern blot analysis of washout experiments comparing the wild-type ChiX to the ChiXΔARN mutant. These experiments were carried out in wild-type and isogenic mutant derivatives of SG30200, each carrying ΔchiX::kan [WT hfq+ (DJS2784), hfqR16A (DJS2786), and hfqY25D (DJS2789)] and harboring a plasmid that expressed the wild-type ChiX (pBR-ChiX) or the ChiXΔARN mutant (pDJS2211). The washouts were carried out similarly to the experiments in Fig2, with the exception that 5 μg of total RNA was analyzed for each sample.
Quantitation of the Northern blot in (B). Quantification was carried out as for Fig EV2. Wild-type ChiX results are shown with a solid line and ChiXΔARN with dotted lines and WT in black, R16A in purple, and Y25D in blue.
β-Galactosidase activity measured in a PBAD-chiP-lacZ ΔchiX::kan strain (DJS2991) carrying a plasmid expressing wild-type ChiX (pBR-ChiX), ChiXΔARN (pDJS2211), or a vector control (pBR-plac). Strains were grown in LB medium containing 100 μg/ml ampicillin, 10 μM IPTG, and 0.002% arabinose at 37°C to early stationary phase (OD600 ∼ 1.0) and assayed for β-galactosidase activity. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
Predicted secondary structure of the wild-type MgrR and the corresponding full ARN deletion mutant (MgrRΔARN) without central predicted stem-loop for MgrR. Color codes are as for Fig4A, with pairing to the eptB target (Moon & Gottesman, 2009) in yellow.
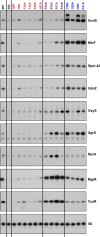
Northern blot analysis of washout experiments comparing wild-type MgrR to the MgrRΔARN mutant. These experiments were carried out in wild-type and isogenic mutant derivatives of SG30200, each carrying ΔmgrR::kan [WT hfq (DJS2963), hfqR16A (DJS2965), and hfqY25D (DJS2966)] and harboring a plasmid that expressed the wild-type MgrR (pBR-MgrR) or the MgrRΔARN mutant (pDJS2225). The washouts were carried out as for Fig2, with the exception that 5 μg of total RNA was analyzed for each sample.
Quantitation of the Northern membrane in (F) as for Fig EV2.
β-galactosidase activity measured in a PBAD-eptB-lacZ ΔmgrR::kan strain (DJS3003) carrying a plasmid expressing wild-type MgrR (pBR-MgrR), MgrRΔARN (pDJS2225), or a vector control (pBR-plac). Samples were treated and analyzed as in (D), except that the LB contained 0.2% arabinose. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
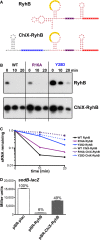
Reported secondary structure of the wild-type RyhB (Geissmann & Touati, 2004) and the predicted secondary structure of the chimera in which the 5′ end of ChiX was fused to truncated RyhB (ChiX-RyhB). Color codes are as for Fig4A, with region of pairing to the sodB target (Geissmann & Touati, 2004) in yellow and the ARN motifs in blue. Red nucleotides correspond to bases from RyhB, and black nucleotides correspond to bases from ChiX.
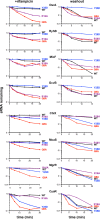
Northern blot analysis of washout experiments to compare the stability of the wild-type RyhB to the ChiX-RyhB chimera. These experiments were carried out in hfq+ ΔchiX::kan (DJS2784), hfqR16A ΔchiX::kan (DJS2786) and hfqY25D ΔchiX::kan (DJS2789), harboring a plasmid that expressed the wild-type RyhB (pBR-RyhB) or the ChiX-RyhB mutant (pDJS2219) under control of a plac promoter. The washouts were carried out as in Fig2, with the exception that 5 μg of total RNA was analyzed for each sample.
Quantitation of the Northern membrane in (B) as in Fig EV2.
β-Galactosidase activity measured in NRD537 (PBAD-sodB-lacZ) carrying a plasmid expressing wild-type RyhB (pBR-RyhB), ChiX-RyhB (pDJS2219), or a vector control (pBR-plac). Samples were treated and analyzed as in Fig4D. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
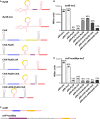
Secondary structures of the wild-type RyhB and the corresponding predicted secondary structures of the various ChiX/RyhB chimeras as in Fig5A.
β-Galactosidase activity measured in NRD537 (PBAD-sodB-lacZ) carrying a plasmid expressing a vector control (pBR-plac), wild-type RyhB (pBR-RyhB), wild-type ChiX (pBR-ChiX), ChiX-RyhB (pDJS2219), RyhBΔUA (pDJS2227), ChiX-RyhBΔUA (pDJS2226), ChiX-RyhB-ChiX (pDJS2229), or ChiXΔARN-RyhB-ChiX (pDJS2230). Samples were treated and analyzed as in Fig4D. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
Sequences from the 5′ UTR of PBAD-sodB-lacZ and PBAD-chiP+sodBbp-lacZ fusions. Color codes show the sequence for the base-pairing region to the mRNA target sodB to the sRNA RyhB highlighted in yellow and the UA element from chiP in purple. Red text represents bases from sodB, and black text represents bases from chiP.
β-Galactosidase activity measured in DJS2985 (PBAD-chiP+sodBbp-lacZ) carrying the same set of plasmids as in (B). Samples were treated and analyzed as in Fig4D. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
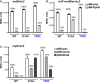
Derivatives of NRD537, each with PBAD-sodB-lacZ [WT hfq (DJS2683), hfqR16A (DJS2686), and hfqY25D (DJS2687)] and carrying a plasmid expressing wild-type RyhB (pBR-RyhB) or a vector control (pBR-plac), were grown and assayed as in Fig4D.
Derivatives of DJS2985, each with PBAD-chiP+sodBbp-lacZ [WT hfq (DJS3009), hfqR16A (DJS3010), and hfqY25D (DJS3011)] and carrying a plasmid expressing wild-type RyhB (pBR-RyhB) or a vector control (pBR-plac). Samples were treated as in Fig4D.
Derivatives of MPK0379, each with PBAD-csgD-lacZ ΔmcaS::kan ΔpgaA::cat [WT hfq (DJS3017), hfqR16A (DJS3018), and hfqY25D (DJS3019)] and carrying a plasmid expressing wild-type McaS (pBR-McaS), wild-type OmrA (pBR-OmrA), or a vector control (pBR-plac), were grown and assayed as in Fig4D except that the LB contained 0.02% arabinose.
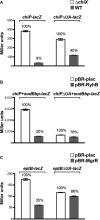
β-Galactosidase activity measured in DJS2979 (PBAD-chiP-lacZ), DJS2982 (PBAD-chiPΔUA-lacZ), and the ΔchiX derivatives (DJS2991 and DJS2994, respectively). Samples were treated as in Fig4D.
β-Galactosidase activity measured in DJS2985 (PBAD-chiPsodBbp-lacZ) or DJS2998 (PBAD-chiPΔUA+sodBbp-lacZ) carrying a plasmid expressing wild-type RyhB (pBR-RyhB) or a vector control (pBR-plac). Samples were treated as in Fig4D.
β-Galactosidase activity measured in DJS3003 (PBAD-eptB-lacZ ΔmgrR::kan) or DJS3004 (PBAD-eptBΔUA –lacZ ΔmgrR::kan) carrying a plasmid expressing wild-type MgrR (pBR-MgrR) or a vector control (pBR-plac). Samples were treated as in Fig4D, except that the LB contained 0.2% arabinose.
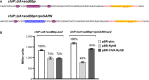
Sequences from the 5′ UTR of PBAD-chiPΔUA+sodBbp-lacZ and PBAD-chiPΔUA+sodBbp+rpoSARN-lacZ fusions. Color codes show the sequence for the base-pairing region to the mRNA target sodB to the sRNA RyhB highlighted in yellow, remaining sequence of the mutated UA element from chiP in purple, and ARN motifs from the 5′ UTR of rpoS in blue. Black text represents bases from chiP, and red text represents bases from sodB.
β-Galactosidase activity measured in DJS2998 (PBAD-chiPΔUA+sodBbp-lacZ) or DJS3015 (PBAD-chiPΔUA+sodBbp+rpoSARN-lacZ) carrying a plasmid expressing wild-type RyhB (pBR-RyhB), ChiX-RyhB (pDJS2219), or a vector control (pBR-plac). Samples were treated as in Fig4D, except that the LB contained 100 μM IPTG.
Similar articles
-
Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells.Elife. 2021 Feb 22;10:e64207. doi: 10.7554/eLife.64207. Elife. 2021. PMID: 33616037 Free PMC article.
-
Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets.J Mol Biol. 2013 Oct 9;425(19):3678-97. doi: 10.1016/j.jmb.2013.01.006. Epub 2013 Jan 11. J Mol Biol. 2013. PMID: 23318956 Free PMC article.
-
The binding of Class II sRNA MgrR to two different sites on matchmaker protein Hfq enables efficient competition for Hfq and annealing to regulated mRNAs.RNA. 2018 Dec;24(12):1761-1784. doi: 10.1261/rna.067777.118. Epub 2018 Sep 14. RNA. 2018. PMID: 30217864 Free PMC article.
-
Structure and RNA-binding properties of the bacterial LSm protein Hfq.RNA Biol. 2013 Apr;10(4):610-8. doi: 10.4161/rna.24201. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535768 Free PMC article. Review.
-
Cycling of RNAs on Hfq.RNA Biol. 2013 Apr;10(4):619-26. doi: 10.4161/rna.24044. Epub 2013 Mar 6. RNA Biol. 2013. PMID: 23466677 Free PMC article. Review.
Cited by
-
Small RNAs OmrA and OmrB promote class III flagellar gene expression by inhibiting the synthesis of anti-Sigma factor FlgM.RNA Biol. 2020 Jun;17(6):872-880. doi: 10.1080/15476286.2020.1733801. Epub 2020 Mar 5. RNA Biol. 2020. PMID: 32133913 Free PMC article.
-
Riboswitch and small RNAs modulate btuB translation initiation in Escherichia coli and trigger distinct mRNA regulatory mechanisms.Nucleic Acids Res. 2024 Jun 10;52(10):5852-5865. doi: 10.1093/nar/gkae347. Nucleic Acids Res. 2024. PMID: 38742638 Free PMC article.
-
An acetyltranferase moonlights as a regulator of the RNA binding repertoire of the RNA chaperone Hfq in Escherichia coli.Proc Natl Acad Sci U S A. 2023 Dec 5;120(49):e2311509120. doi: 10.1073/pnas.2311509120. Epub 2023 Nov 27. Proc Natl Acad Sci U S A. 2023. PMID: 38011569 Free PMC article.
-
Control of iron acquisition by multiple small RNAs unravels a new role for transcriptional terminator loops in gene regulation.Nucleic Acids Res. 2024 Dec 11;52(22):13775-13791. doi: 10.1093/nar/gkae1131. Nucleic Acids Res. 2024. PMID: 39611574 Free PMC article.
-
The Escherichia Coli Hfq Protein: An Unattended DNA-Transactions Regulator.Front Mol Biosci. 2016 Jul 28;3:36. doi: 10.3389/fmolb.2016.00036. eCollection 2016. Front Mol Biosci. 2016. PMID: 27517037 Free PMC article. Review.
References
-
- Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol. 2010;78:380–394. - PubMed
-
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

