Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma
- PMID: 26374103
- PMCID: PMC4570720
- DOI: 10.1186/s40246-015-0044-0
Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma
Abstract
Background: Esophageal adenocarcinoma (EA) is among the leading causes of cancer mortality, especially in developed countries. A high level of somatic copy number alterations (CNAs) accumulates over the decades in the progression from Barrett's esophagus, the precursor lesion, to EA. Accurate identification of somatic CNAs is essential to understand cancer development. Many studies have been conducted for the detection of CNA in EA using microarrays. Next-generation sequencing (NGS) technologies are believed to have advantages in sensitivity and accuracy to detect CNA, yet no NGS-based CNA detection in EA has been reported.
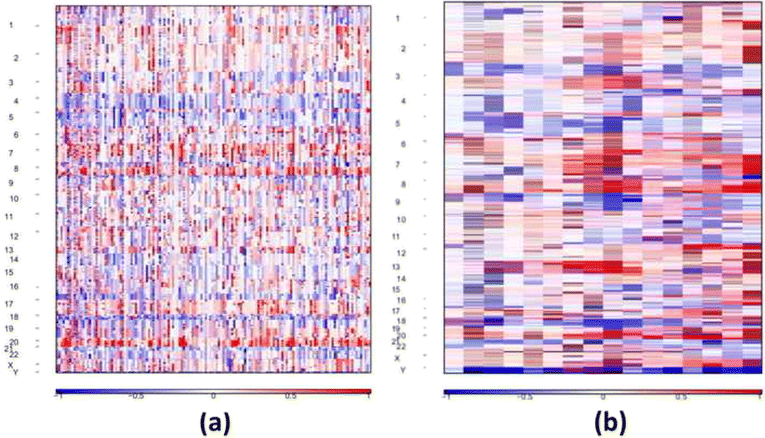
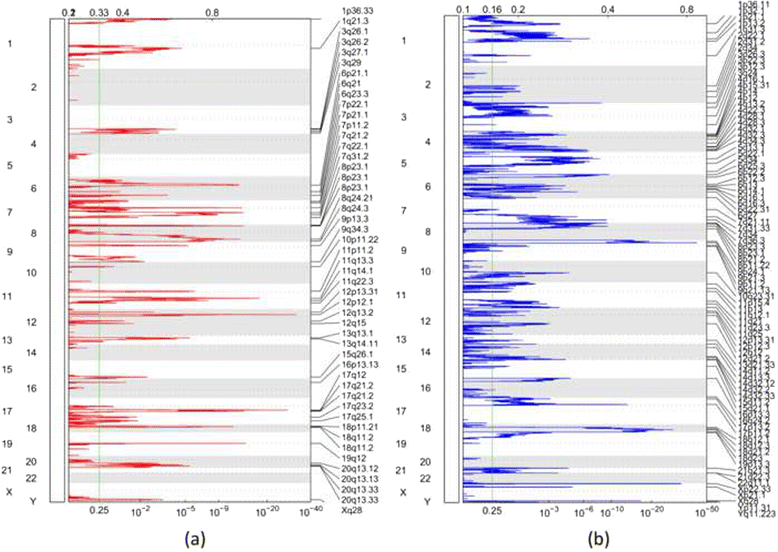
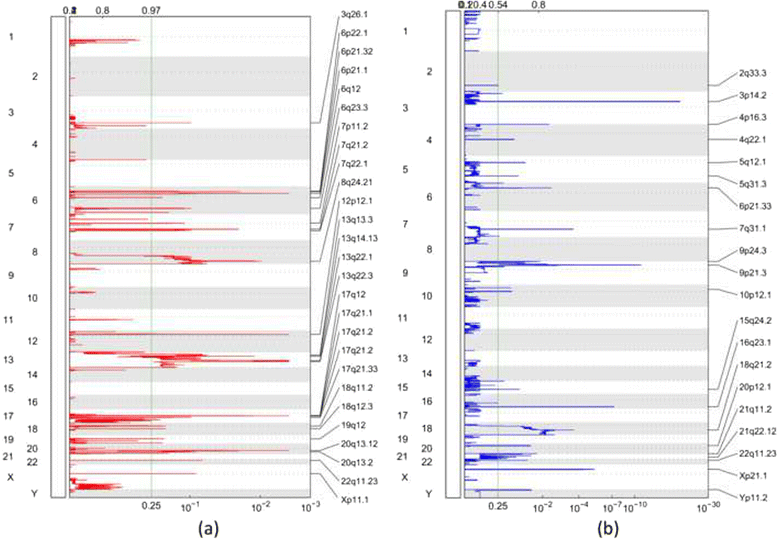
Results: In this study, we analyzed whole-exome (WES) and whole-genome sequencing (WGS) data for detecting CNA from a published large-scale genomic study of EA. Two specific comparisons were conducted. First, the recurrent CNAs based on WGS and WES data from 145 EA samples were compared to those found in five previous microarray-based studies. We found that the majority of the previously identified regions were also detected in this study. Interestingly, some novel amplifications and deletions were discovered using the NGS data. In particular, SKI and PRKCZ detected in a deletion region are involved in transforming growth factor-β pathway, suggesting the potential utility of novel biomarkers for EA. Second, we compared CNAs detected in WGS and WES data from the same 15 EA samples. No large-scale CNA was identified statistically more frequently by WES or WGS, while more focal-scale CNAs were detected by WGS than by WES.
Conclusions: Our results suggest that NGS can replace microarrays to detect CNA in EA. WGS is superior to WES in that it can offer finer resolution for the detection, though if the interest is on recurrent CNAs, WES can be preferable to WGS for its cost-effectiveness.
Figures
Similar articles
-
Chromosomal instability and copy number alterations in Barrett's esophagus and esophageal adenocarcinoma.Clin Cancer Res. 2009 May 15;15(10):3305-14. doi: 10.1158/1078-0432.CCR-08-2494. Epub 2009 May 5. Clin Cancer Res. 2009. PMID: 19417022 Free PMC article.
-
Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants.Proc Natl Acad Sci U S A. 2015 Apr 28;112(17):5473-8. doi: 10.1073/pnas.1418631112. Epub 2015 Mar 31. Proc Natl Acad Sci U S A. 2015. PMID: 25827230 Free PMC article.
-
An evaluation of copy number variation detection tools for cancer using whole exome sequencing data.BMC Bioinformatics. 2017 May 31;18(1):286. doi: 10.1186/s12859-017-1705-x. BMC Bioinformatics. 2017. PMID: 28569140 Free PMC article.
-
The Evolving Genomic Landscape of Barrett's Esophagus and Esophageal Adenocarcinoma.Gastroenterology. 2017 Sep;153(3):657-673.e1. doi: 10.1053/j.gastro.2017.07.007. Epub 2017 Jul 14. Gastroenterology. 2017. PMID: 28716721 Free PMC article. Review.
-
Free-access copy-number variant detection tools for targeted next-generation sequencing data.Mutat Res Rev Mutat Res. 2019 Jan-Mar;779:114-125. doi: 10.1016/j.mrrev.2019.02.005. Epub 2019 Feb 23. Mutat Res Rev Mutat Res. 2019. PMID: 31097148 Review.
Cited by
-
Correlation between NGS panel-based mutation results and clinical information in colorectal cancer patients.Heliyon. 2024 Apr 5;10(7):e29299. doi: 10.1016/j.heliyon.2024.e29299. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38623252 Free PMC article.
-
Stratification of clear cell renal cell carcinoma (ccRCC) genomes by gene-directed copy number alteration (CNA) analysis.PLoS One. 2017 May 9;12(5):e0176659. doi: 10.1371/journal.pone.0176659. eCollection 2017. PLoS One. 2017. PMID: 28486536 Free PMC article.
-
The Ubiquitin-Conjugating Enzyme E2 O (UBE2O) and Its Therapeutic Potential in Human Leukemias and Solid Tumors.Cancers (Basel). 2024 Sep 3;16(17):3064. doi: 10.3390/cancers16173064. Cancers (Basel). 2024. PMID: 39272922 Free PMC article. Review.
-
High-resolution genomic profiling and locus-specific FISH in subcutaneous and visceral adipose tissue of obese patients.Front Genet. 2024 Mar 7;14:1323052. doi: 10.3389/fgene.2023.1323052. eCollection 2023. Front Genet. 2024. PMID: 38516060 Free PMC article.
-
Whole-genome Sequencing of Follicular Thyroid Carcinomas Reveal Recurrent Mutations in MicroRNA Processing Subunit DGCR8.J Clin Endocrinol Metab. 2021 Oct 21;106(11):3265-3282. doi: 10.1210/clinem/dgab471. J Clin Endocrinol Metab. 2021. PMID: 34171097 Free PMC article.
References
-
- Greshock J, Feng B, Nogueira C, Ivanova E, Perna I, Nathanson K, et al. A comparison of DNA copy number profiling platforms. Cancer Res. 2007;67(21):10173–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

