Ligand-dependent genomic function of glucocorticoid receptor in triple-negative breast cancer
- PMID: 26374485
- PMCID: PMC4573460
- DOI: 10.1038/ncomms9323
Ligand-dependent genomic function of glucocorticoid receptor in triple-negative breast cancer
Abstract
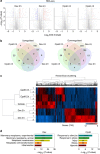
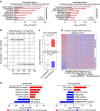
Glucocorticoids (GCs) have been widely used as coadjuvants in the treatment of solid tumours, but GC treatment may be associated with poor pharmacotherapeutic response or prognosis. The genomic action of GC in these tumours is largely unknown. Here we find that dexamethasone (Dex, a synthetic GC)-regulated genes in triple-negative breast cancer (TNBC) cells are associated with drug resistance. Importantly, these GC-regulated genes are aberrantly expressed in TNBC patients and are associated with unfavourable clinical outcomes. Interestingly, in TNBC cells, Compound A (CpdA, a selective GR modulator) only regulates a small number of genes not involved in carcinogenesis and therapy resistance. Mechanistic studies using a ChIP-exo approach reveal that Dex- but not CpdA-liganded glucocorticoid receptor (GR) binds to a single glucocorticoid response element (GRE), which drives the expression of pro-tumorigenic genes. Our data suggest that development of safe coadjuvant therapy should consider the distinct genomic function between Dex- and CpdA-liganded GR.
Figures


References
-
- Schmidt S. et al. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death and Differ. 11, (Suppl 1): S45–S55 (2004). - PubMed
-
- Rutz H. P. Effects of corticosteroid use on treatment of solid tumours. Lancet 360, 1969–1970 (2002). - PubMed
-
- Zhang C. et al. Clinical and mechanistic aspects of glucocorticoid-induced chemotherapy resistance in the majority of solid tumors. Cancer Biol. Ther. 6, 278–287 (2007). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

