Genetic dissection of the Down syndrome critical region
- PMID: 26374847
- PMCID: PMC4614710
- DOI: 10.1093/hmg/ddv364
Genetic dissection of the Down syndrome critical region
Abstract
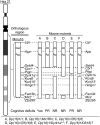
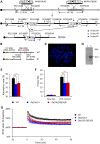
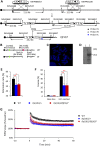
Down syndrome (DS), caused by trisomy 21, is the most common chromosomal disorder associated with developmental cognitive deficits. Despite intensive efforts, the genetic mechanisms underlying developmental cognitive deficits remain poorly understood, and no treatment has been proven effective. The previous mouse-based experiments suggest that the so-called Down syndrome critical region of human chromosome 21 is an important region for this phenotype, which is demarcated by Setd4/Cbr1 and Fam3b/Mx2. We first confirmed the importance of the Cbr1-Fam3b region using compound mutant mice, which carry a duplication spanning the entire human chromosome 21 orthologous region on mouse chromosome 16 [Dp(16)1Yey] and Ms1Rhr. By dividing the Setd4-Mx2 region into complementary Setd4-Kcnj6 and Kcnj15-Mx2 intervals, we started an unbiased dissection through generating and analyzing Dp(16)1Yey/Df(16Setd4-Kcnj6)Yey and Dp(16)1Yey/Df(16Kcnj15-Mx2)Yey mice. Surprisingly, the Dp(16)1Yey-associated cognitive phenotypes were not rescued by either deletion in the compound mutants, suggesting the possible presence of at least one causative gene in each of the two regions. The partial rescue by a Dyrk1a mutation in a compound mutant carrying Dp(16)1Yey and the Dyrk1a mutation confirmed the causative role of Dyrk1a, whereas the absence of a similar rescue by Df(16Dyrk1a-Kcnj6)Yey in Dp(16)1Yey/Df(16Dyrk1a-Kcnj6)Yey mice demonstrated the importance of Kcnj6. Our results revealed the high levels of complexities of gene actions and interactions associated with the Setd4/Cbr1-Fam3b/Mx2 region as well as their relationship with developmental cognitive deficits in DS.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Antonarakis S.E., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S. (2004) Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat. Rev. Genet., 5, 725–738. - PubMed
-
- Roizen N.J., Patterson D. (2003) Down's syndrome. Lancet, 361, 1281–1289. - PubMed
-
- Parker S.E., Mai C.T., Canfield M.A., Rickard R., Wang Y., Meyer R.E., Anderson P., Mason C.A., Collins J.S., Kirby R.S. et al. (2010) Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res. A Clin. Mol. Teratol., 88, 1008–1016. - PubMed
-
- Natoli J.L., Ackerman D.L., McDermott S., Edwards J.G. (2012) Prenatal diagnosis of Down syndrome: a systematic review of termination rates (1995–2011). Prenat. Diagn., 32, 142–153. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

