Assembling the puzzle: Oligomerization of α-pore forming proteins in membranes
- PMID: 26375417
- PMCID: PMC4869852
- DOI: 10.1016/j.bbamem.2015.09.013
Assembling the puzzle: Oligomerization of α-pore forming proteins in membranes
Abstract
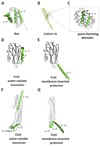
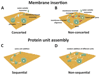
Pore forming proteins (PFPs) share the ability of creating pores that allow the passage of ions, proteins or other constituents through a wide variety of target membranes, ranging from bacteria to humans. They often cause cell death, as pore formation disrupts the membrane permeability barrier required for maintaining cell homeostasis. The organization into supramolecular complexes or oligomers that pierce the membrane is a common feature of PFPs. However, the molecular pathway of self-assembly and pore opening remains unclear. Here, we review the most recent discoveries in the mechanism of membrane oligomerization and pore formation of a subset of PFPs, the α-PFPs, whose pore-forming domains are formed by helical segments. Only now we are starting to grasp the molecular details of their function, mainly thanks to the introduction of single molecule microscopy and nanoscopy techniques. This article is part of a Special Issue entitled: Pore-forming toxins edited by Mauro Dalla Serra and Franco Gambale.
Keywords: Membrane; Pore forming proteins (PFPs); Pore forming toxins (PFTs); Pore structure; Protein oligomerization.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Iacovache I, Bischofberger M, van der Goot FG. Structure and assembly of pore-forming proteins. Curr Opin Struct Biol. 2010;20:241–246. - PubMed
-
- Bischofberger M, Iacovache I, Gisou van der Goot F. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe. 2012;12:266–275. - PubMed
-
- Cosentino K, García-Sáez AJ. Mitochondrial alterations in apoptosis. Chem Phys Lipids. 2014;181:62–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

