Automated tracking of tumor-stroma morphology in microtissues identifies functional targets within the tumor microenvironment for therapeutic intervention
- PMID: 26375443
- PMCID: PMC4745780
- DOI: 10.18632/oncotarget.5046
Automated tracking of tumor-stroma morphology in microtissues identifies functional targets within the tumor microenvironment for therapeutic intervention
Abstract
Cancer-associated fibroblasts (CAFs) constitute an important part of the tumor microenvironment and promote invasion via paracrine functions and physical impact on the tumor. Although the importance of including CAFs into three-dimensional (3D) cell cultures has been acknowledged, computational support for quantitative live-cell measurements of complex cell cultures has been lacking. Here, we have developed a novel automated pipeline to model tumor-stroma interplay, track motility and quantify morphological changes of 3D co-cultures, in real-time live-cell settings. The platform consists of microtissues from prostate cancer cells, combined with CAFs in extracellular matrix that allows biochemical perturbation. Tracking of fibroblast dynamics revealed that CAFs guided the way for tumor cells to invade and increased the growth and invasiveness of tumor organoids. We utilized the platform to determine the efficacy of inhibitors in prostate cancer and the associated tumor microenvironment as a functional unit. Interestingly, certain inhibitors selectively disrupted tumor-CAF interactions, e.g. focal adhesion kinase (FAK) inhibitors specifically blocked tumor growth and invasion concurrently with fibroblast spreading and motility. This complex phenotype was not detected in other standard in vitro models. These results highlight the advantage of our approach, which recapitulates tumor histology and can significantly improve cancer target validation in vitro.
Keywords: 3D co-culture; cancer associated fibroblast (CAF); focal adhesion kinase (FAK); invasion; phenotypic screening.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures
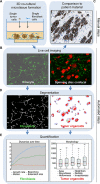
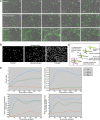
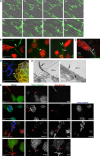
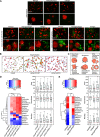


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

