Extremely High Mutation Rate of HIV-1 In Vivo
- PMID: 26375597
- PMCID: PMC4574155
- DOI: 10.1371/journal.pbio.1002251
Extremely High Mutation Rate of HIV-1 In Vivo
Abstract
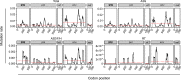
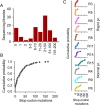
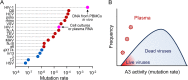
Rates of spontaneous mutation critically determine the genetic diversity and evolution of RNA viruses. Although these rates have been characterized in vitro and in cell culture models, they have seldom been determined in vivo for human viruses. Here, we use the intrapatient frequency of premature stop codons to quantify the HIV-1 genome-wide rate of spontaneous mutation in DNA sequences from peripheral blood mononuclear cells. This reveals an extremely high mutation rate of (4.1 ± 1.7) × 10-3 per base per cell, the highest reported for any biological entity. Sequencing of plasma-derived sequences yielded a mutation frequency 44 times lower, indicating that a large fraction of viral genomes are lethally mutated and fail to reach plasma. We show that the HIV-1 reverse transcriptase contributes only 2% of mutations, whereas 98% result from editing by host cytidine deaminases of the A3 family. Hypermutated viral sequences are less abundant in patients showing rapid disease progression compared to normal progressors, highlighting the antiviral role of A3 proteins. However, the amount of A3-mediated editing varies broadly, and we find that low-edited sequences are more abundant among rapid progressors, suggesting that suboptimal A3 activity might enhance HIV-1 genetic diversity and pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Cellular Defense Enzyme Drives Exceptionally High Rate of Mutation in HIV.PLoS Biol. 2015 Sep 16;13(9):e1002252. doi: 10.1371/journal.pbio.1002252. eCollection 2015 Sep. PLoS Biol. 2015. PMID: 26375682 Free PMC article.
References
-
- Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002;2:28–36. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

