Identification of a Novel Human Papillomavirus, Type HPV199, Isolated from a Nasopharynx and Anal Canal, and Complete Genomic Characterization of Papillomavirus Species Gamma-12
- PMID: 26375679
- PMCID: PMC4574437
- DOI: 10.1371/journal.pone.0138628
Identification of a Novel Human Papillomavirus, Type HPV199, Isolated from a Nasopharynx and Anal Canal, and Complete Genomic Characterization of Papillomavirus Species Gamma-12
Abstract
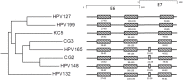
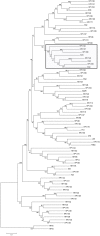
The novel human papillomavirus type 199 (HPV199) was initially identified in a nasopharyngeal swab sample obtained from a 25 year-old immunocompetent male. The complete genome of HPV199 is 7,184 bp in length with a GC content of 36.5%. Comparative genomic characterization of HPV199 and its closest relatives showed the classical genomic organization of Gammapapillomaviruses (Gamma-PVs). HPV199 has seven major open reading frames (ORFs), encoding five early (E1, E2, E4, E6, and E7) and two late (L1 and L2) proteins, while lacking the E5 ORF. The long control region (LCR) of 513 bp is located between the L1 and E6 ORFs. Phylogenetic analysis additionally confirmed that HPV-199 clusters into the Gamma-PV genus, species Gamma-12, additionally containing HPV127, HV132, HPV148, HPV165, and three putative HPV types: KC5, CG2 and CG3. HPV199 is most closely related to HPV127 (nucleotide identity 77%). The complete viral genome sequence of additional HPV199 isolate was determined from anal canal swab sample. Two HPV199 complete viral sequences exhibit 99.4% nucleotide identity. To the best of our knowledge, this is the first member of Gamma-PV with complete nucleotide sequences determined from two independent clinical samples. To evaluate the tissue tropism of the novel HPV type, 916 clinical samples were tested using HPV199 type-specific real-time PCR: HPV199 was detected in 2/76 tissue samples of histologically confirmed common warts, 2/108 samples of eyebrow hair follicles, 2/137 anal canal swabs obtained from individuals with clinically evident anal pathology, 4/184 nasopharyngeal swabs and 3/411 cervical swabs obtained from women with normal cervical cytology. Although HPV199 was found in 1.4% of cutaneous and mucosal samples only, it exhibits dual tissue tropism. According to the results of our study and literature data, dual tropism of all Gamma-12 members is highly possible.
Conflict of interest statement
Figures

Similar articles
-
Identification of a novel human papillomavirus by metagenomic analysis of vaginal swab samples from pregnant women.Virol J. 2016 Jul 4;13:122. doi: 10.1186/s12985-016-0583-6. Virol J. 2016. PMID: 27411548 Free PMC article.
-
Characterization of novel human papillomavirus types 157, 158 and 205 from healthy skin and recombination analysis in genus γ-Papillomavirus.Infect Genet Evol. 2016 Aug;42:20-9. doi: 10.1016/j.meegid.2016.04.018. Epub 2016 Apr 21. Infect Genet Evol. 2016. PMID: 27108808
-
Characterization of two novel gammapapillomaviruses, HPV179 and HPV184, isolated from common warts of a renal-transplant recipient.PLoS One. 2015 Mar 6;10(3):e0119154. doi: 10.1371/journal.pone.0119154. eCollection 2015. PLoS One. 2015. PMID: 25748516 Free PMC article.
-
[Genomic organization and proteins of human papillomavirus].Mikrobiyol Bul. 2012 Jul;46(3):507-15. Mikrobiyol Bul. 2012. PMID: 22951665 Review. Turkish.
-
[Virological and carcinogenic aspects of HPV].Bull Acad Natl Med. 2007 Mar;191(3):611-23; discussion 623. Bull Acad Natl Med. 2007. PMID: 18072657 Review. French.
Cited by
-
HPV Infections-Classification, Pathogenesis, and Potential New Therapies.Int J Mol Sci. 2024 Jul 11;25(14):7616. doi: 10.3390/ijms25147616. Int J Mol Sci. 2024. PMID: 39062859 Free PMC article. Review.
-
Mouse papillomavirus infections spread to cutaneous sites with progression to malignancy.J Gen Virol. 2017 Oct;98(10):2520-2529. doi: 10.1099/jgv.0.000926. J Gen Virol. 2017. PMID: 28942760 Free PMC article.
-
Mucosal and Cutaneous Human Papillomavirus Infections and Cancer Biology.Front Oncol. 2019 May 8;9:355. doi: 10.3389/fonc.2019.00355. eCollection 2019. Front Oncol. 2019. PMID: 31134154 Free PMC article. Review.
-
The genetic diversity of human papillomavirus types from the species Gammapapillomavirus 15: HPV135, HPV146, and HPV179.PLoS One. 2021 May 6;16(5):e0249829. doi: 10.1371/journal.pone.0249829. eCollection 2021. PLoS One. 2021. PMID: 33956809 Free PMC article.
-
Discovery, characterisation and genomic variation of six novel Gammapapillomavirus types from penile swabs in South Africa.Papillomavirus Res. 2019 Jun;7:102-111. doi: 10.1016/j.pvr.2019.02.005. Epub 2019 Mar 4. Papillomavirus Res. 2019. PMID: 30844514 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

