High Prevalence and Clinical Relevance of Genes Affected by Chromosomal Breaks in Colorectal Cancer
- PMID: 26375816
- PMCID: PMC4574474
- DOI: 10.1371/journal.pone.0138141
High Prevalence and Clinical Relevance of Genes Affected by Chromosomal Breaks in Colorectal Cancer
Abstract
Background: Cancer is caused by somatic DNA alterations such as gene point mutations, DNA copy number aberrations (CNA) and structural variants (SVs). Genome-wide analyses of SVs in large sample series with well-documented clinical information are still scarce. Consequently, the impact of SVs on carcinogenesis and patient outcome remains poorly understood. This study aimed to perform a systematic analysis of genes that are affected by CNA-associated chromosomal breaks in colorectal cancer (CRC) and to determine the clinical relevance of recurrent breakpoint genes.
Methods: Primary CRC samples of patients with metastatic disease from CAIRO and CAIRO2 clinical trials were previously characterized by array-comparative genomic hybridization. These data were now used to determine the prevalence of CNA-associated chromosomal breaks within genes across 352 CRC samples. In addition, mutation status of the commonly affected APC, TP53, KRAS, PIK3CA, FBXW7, SMAD4, BRAF and NRAS genes was determined for 204 CRC samples by targeted massive parallel sequencing. Clinical relevance was assessed upon stratification of patients based on gene mutations and gene breakpoints that were observed in >3% of CRC cases.
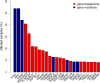
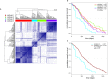
Results: In total, 748 genes were identified that were recurrently affected by chromosomal breaks (FDR <0.1). MACROD2 was affected in 41% of CRC samples and another 169 genes showed breakpoints in >3% of cases, indicating that prevalence of gene breakpoints is comparable to the prevalence of well-known gene point mutations. Patient stratification based on gene breakpoints and point mutations revealed one CRC subtype with very poor prognosis.
Conclusions: We conclude that CNA-associated chromosomal breaks within genes represent a highly prevalent and clinically relevant subset of SVs in CRC.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

