A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers
- PMID: 26376692
- PMCID: PMC4574073
- DOI: 10.1186/s12864-015-1902-9
A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers
Abstract
Background: Colorectal cancer is a major contributor to cancer morbidity and mortality. Tandem repeat instability and its effect on cancer phenotypes remain so far poorly studied on a genome-wide scale.
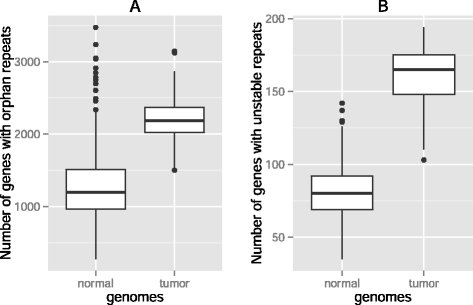
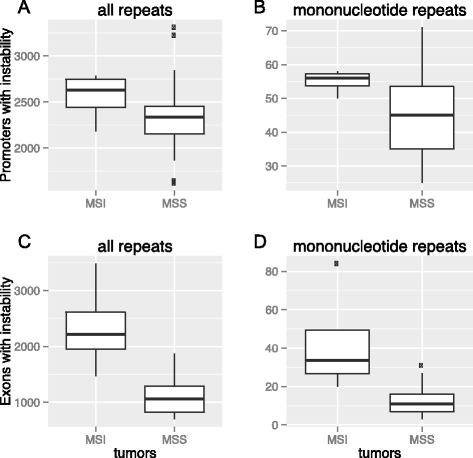
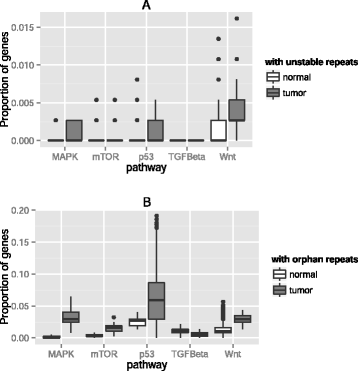
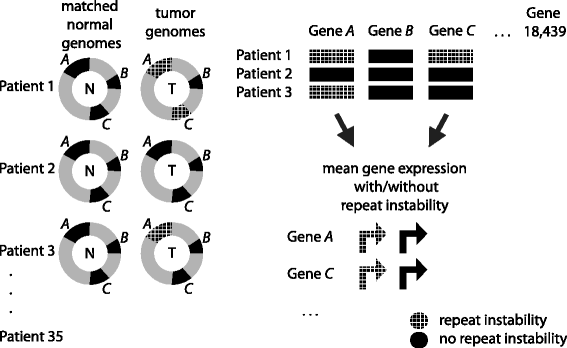
Results: Here we analyze the genomes of 35 colorectal tumors and their matched normal (healthy) tissues for two types of tandem repeat instability, de-novo repeat gain or loss and repeat copy number variation. Specifically, we study for the first time genome-wide repeat instability in the promoters and exons of 18,439 genes, and examine the association of repeat instability with genome-scale gene expression levels. We find that tumors with a microsatellite instable (MSI) phenotype are enriched in genes with repeat instability, and that tumor genomes have significantly more genes with repeat instability compared to healthy tissues. Genes in tumor genomes with repeat instability in their promoters are significantly less expressed and show slightly higher levels of methylation. Genes in well-studied cancer-associated signaling pathways also contain significantly more unstable repeats in tumor genomes. Genes with such unstable repeats in the tumor-suppressor p53 pathway have lower expression levels, whereas genes with repeat instability in the MAPK and Wnt signaling pathways are expressed at higher levels, consistent with the oncogenic role they play in cancer.
Conclusions: Our results suggest that repeat instability in gene promoters and associated differential gene expression may play an important role in colorectal tumors, which is a first step towards the development of more effective molecular diagnostic approaches centered on repeat instability.
Figures




References
-
- Gemayel R, Vinces MD. Legendre M. Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences. Annu Rev Genet: Verstrepen KJ; 2010. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

